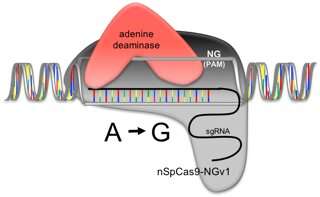
Adenine in the target sequence is substituted by guanine. Credit: Ehime University
Researchers have developed a new genome editing technology for rice, combining adenine-to-guanine single-base editing technology and Cas9 with an extended targeting scope. They report that it is possible to efficiently introduce base substitution mutations in rice genes and plan to expand the research to citrus fruit breeding.
Clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated protein 9 (Cas9) is a powerful genome editing tool. Cas9 can modify any desired target sequence, but is limited to the sequence just upstream of the protospacer adjacent motif (PAM). SpCas9 derived from Streptococcus pyogenes is the most widely used.
The PAM sequence of SpCas9 is NGG (N is any base and G is guanine) and is the smallest of Cas9s known.
Recently, a new type of SpCas9-NG that reduces this restriction of PAM to NG has been reported. Cas9 usually causes deletions and insertion mutations in genome. The frequency of substitution mutations caused by Cas9 is very low, and even if it occurs, the substitution pattern is random.
Another new technology has been developed that can replace A (adenine) to G (guanine) at the targeted sequence. The researchers in the current study fused these two latest technologies and examined whether the resulting method could replace A-to-G at a desired position in the rice genome. They successfully induced such A-to-G base substitutions in the rice genome. They plan to verify this technology and apply it to citrus, which is the most important crop of Ehime Prefecture, which could lead to the development of new varieties.
More information: Katsuya Negishi et al. An adenine base editor with expanded targeting scope using SpCas9‐ NG v1 in rice, Plant Biotechnology Journal (2019). DOI: 10.1111/pbi.13120
Journal information: Plant Biotechnology Journal
Provided by Ehime University