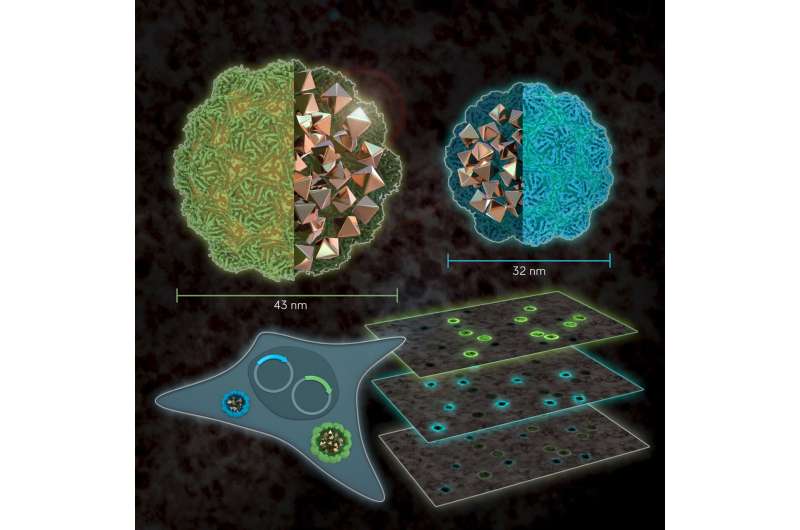
Size matters: EMcapsulins as genetically expressed and multiplexed gene reporters for Electron Microscopy. Credit: Barth van Rossum
Researchers at Helmholtz Zentrum Muenchen have developed a method to visualize gene expression of cells with an electron microscope. Although electron microscopy currently provides the most detailed look into cells, it cannot differentiate which genetic programs run inside individual cells. The new method can now have a closer look by using genetically programmed nanospheres of different sizes as "multicolor" markers, which could even be helpful to investigate how memories are stored in neuronal networks.
What exactly is going on in cells? This question has kept scientists busy for decades. To label small structures, scientists have been using fluorescent proteins. This method works well but has disadvantages due to the relatively poor resolution of light microscopes. Although electron microscopes allow a closer look, says Prof. Dr. Gil Gregor Westmeyer, "so far there are hardly any solutions for multi-color genetic labeling of cells for this technology, such that one can directly tell different cells apart." He leads a research group at the Institute for Biological and Medical Imaging (IBMI) of Helmholtz Zentrum München and is Professor of Molecular Imaging at TUM School of Medicine.
Nanocompartments as multi-color labels for electron microscopy
Westmeyer and colleagues have been working with so-called encapsulins for some time. These are small, non-toxic proteins from bacteria. Encapsulins automatically assemble to nanocompartments in which chemical reactions can run without disturbing the metabolism of the cell. Depending on the experimental conditions, nanocompartments with different diameters are formed within living cells via genetic programming. "Analogous to the palette of colors in fluorescence microscopy, our method turns geometry into a label for electron microscopy," adds Felix Sigmund from Westmeyer's research group.
To achieve strong contrast in the images from the electron microscopy, the researchers use the enzyme ferroxidase, which can be encapsulated in the interior of encapsulins. If iron ions enter the interior lumen through pores of the nanocompartments, divalent iron ions are oxidized by the enzyme into their trivalent form. This creates insoluble iron oxides that remain inside. Metals create good contrasts because they "swallow" electrons—comparable to dense bones in an X-ray image, which strongly absorb X-rays. This special material property of encapsulins makes them clearly visible in the images.
Following neuronal tracts
With their new method, the researchers will now also investigate neural circuits. Despite the impressive resolution of electron microscopy, the method cannot reliably distinguish certain types of neurons within the brain. "With our new reporter genes, we could label specific cells and then read out which type of nerve cell makes which connections and which state the cells are in," adds Westmeyer.
This new reporter technology could thus also help to uncover the exact wiring diagram of brains and to investigate closer how memories are stored in neuronal networks.
More information: Felix Sigmund et al. Iron-Sequestering Nanocompartments as Multiplexed Electron Microscopy Gene Reporters, ACS Nano (2019). DOI: 10.1021/acsnano.9b03140
Felix Sigmund et al. Bacterial encapsulins as orthogonal compartments for mammalian cell engineering, Nature Communications (2018). DOI: 10.1038/s41467-018-04227-3
Journal information: ACS Nano , Nature Communications
Provided by Helmholtz Zentrum München