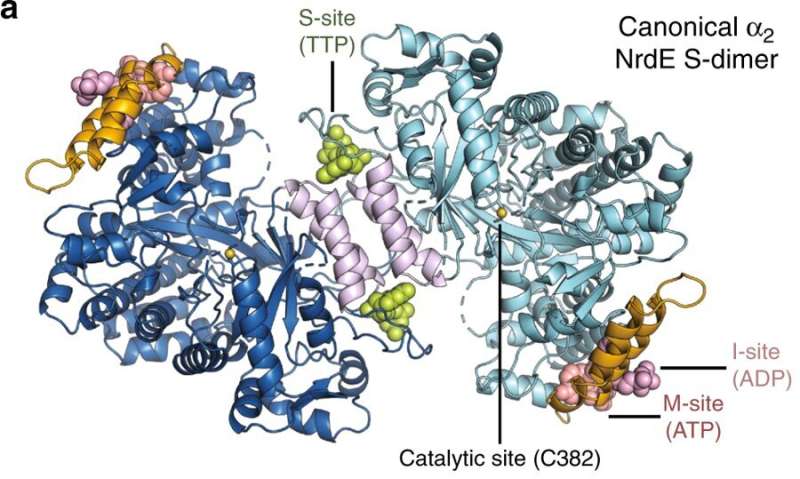
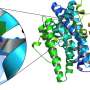
Allosteric sites of the Bacillus subtilis class Ib ribonucleotide reductase. a A 2.50 Å crystal structure of B. subtilis NrdE (α subunit) obtained under activating conditions depicts an S-shaped dimer (“S-dimer”) interfacing at the “specificity” or S-site (lavender). A specificity effector TTP (green) is bound to the S-site, and activating nucleotides, ADP (pink) and ATP (salmon), are bound to two allosteric sites that evolved near the N-terminus of B. subtilis NrdE. A catalytically essential radical is generated at a central cysteine in the catalytic site, C382 (yellow sphere). b B. subtilis NrdF (β subunit) is dimeric and utilizes a dimanganic tyrosyl cofactor (purple spheres) to initiate radical chemistry (PDB: 4DR0)21. A disordered region of the NrdF C-terminus (black dotted lines) is critical for radical transfer. c A recent structure of B. subtilis NrdE co-crystallized with dAMP (purple) depicts a partially inhibited, non-canonical “I-dimer” with the interface formed by the truncated ATP-cone (orange) (PDB: 6CGL)31. d In class Ia RNRs, ATP or dATP binds to the “activity” or A-site in the ATP-cone domain (orange) to mediate changes in quaternary structure and tune overall activity (PDB: 3R1R)20. e Class Ib RNRs only contain partial ATP-cones (orange). B. subtilis NrdE is unusual in that it displays activity regulation and binds dAMP (purple) in the N-terminally located I-site (PDB: 6CGL). f The partial N-terminal cone of class Ib RNRs (top) is structurally homologous to the last two helices of the canonical ATP-cone found in many class Ia RNRs (bottom) but lacks A-site residues. Credit: Nature Communications (2019). DOI: 10.1038/s41467-019-10568-4
New research from Cornell University offers a new pathway for targeting pathogens in the fight against antibiotic resistant bacteria.
As antibiotic resistance rises, the search for new antibiotic strategies has become imperative. Researchers used the Cornell High Energy Synchotron Source (CHESS) to reveal an unexpected mechanism of activation and inactivation in the protein ribonucleotide reductase (RNR).
The findings were published in "Convergent Allostery in Ribonucleotide Reductase" in Nature Communications.
Understanding the "switch" that can turn RNR off provides a possible means to shut off the reproduction of harmful bacteria.
RNRs take ribonucleotides, the building blocks of RNA, and convert them to deoxyribonucleotides, the building blocks of DNA. In all organisms, the regulation of RNRs involves complex mechanisms. Without these mechanisms, DNA replication becomes error-prone, and dangerous mutations could occur.
"Without the RNR enzyme, DNA-based life as we know it could not exist," said first author William Thomas, a graduate student in chemistry and chemical biology. "If we understand the RNR 'off switch' well enough, we can take advantage of it by developing our own ways to toggle it with new antibiotic drug molecules."
This research reveals evolution in action, according to Nozomi Ando, assistant professor of chemistry and the paper's senior author. The lack of the normal regulatory "switch" mechanism may provide an evolutionary advantage for the bacteria they studied.
"Usually the increased chance of mutations is a problem for bacteria, but maybe under certain circumstances it's actually advantageous for an organism to mutate and possibly become resistant to an antibiotic or another stressful situation," she said.
RNRs are not easy proteins to work with or understand, and the researchers said characterizing them in the traditional way has been challenging.
"The combination of small-angle X-ray scattering using CHESS, crystallography, and cryo-electron microscopy is what made this study possible," Ando said.
More information: William C. Thomas et al. Convergent allostery in ribonucleotide reductase, Nature Communications (2019). DOI: 10.1038/s41467-019-10568-4
Journal information: Nature Communications
Provided by Cornell University