Reading the dark heart of chromosomes
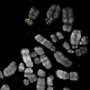
Although the genomes of thousands of plant and animal species have been sequenced, for most of these genomes a significant portion is missing—the highly repetitive DNA. In the midst of these mysterious genome compartments are the centromeres—essential chromosomal regions that allow cells to accurately pass on chromosomes when the cells divide. A new study publishing May 14 in the open-access journal PLOS Biology by the Mellone lab at the University of Connecticut and the Larracuente lab at the University of Rochester combine cutting-edge sequencing technology with molecular and high-resolution microscopy methods to discover the sequences of all centromeres in the fruit fly Drosophila melanogaster, a powerful model organism widely used in biomedical research.
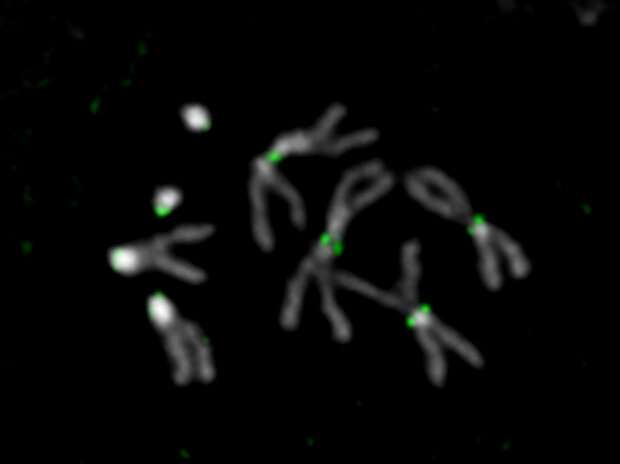
Centromeres have been visible under the microscope for over a century, but little is known about their organization at the DNA level because of the difficulty of accessing highly repetitive DNA with traditional sequencing technologies. To sequence a genome, researchers fragment DNA into 'readable' units and then assemble these units back into contiguous sequences that represent the genome, in a computational process akin to assembling a jigsaw puzzle. While this process works well for unique DNA sequences in and around genes, in repetitive regions of the genome all of the puzzle pieces look identical, making it difficult to figure out how they fit together.
To get around this problem, the authors combined this traditional 'jigsaw' approach with a suite of other methods: sequencing longer sections of DNA, purifying segments of the centromere that stick to a centromere-specific histone protein, and imaging chromatin fibers with high-resolution microscopy. Using these approaches, the authors were able to generate a complete and intact picture of the fly's centromeres, finding that buried within a sea of highly repetitive sequences, there were 'islands' of more complex DNA sequences that might hold the key to how the centromeres function to segregate chromosomes faithfully.
The researchers found that centromeres contain a surprisingly high number of transposable elements—sequences that jump around and selfishly proliferate throughout the genomes. "What is exciting is that the centromere islands are rich in a type of transposable element called retroelements, which we usually consider to be genome parasites," says Professor Amanda Larracuente, co-lead author on the study. A particular retroelement, called G2/Jockey-3, was found in all centromeres, not only in this species of fruit fly, but also in a closely related one, Drosophila simulans.
Their findings suggest that these selfish DNA elements may have a role in centromere function across a wide range of species, as retroelements have been found to be associated with centromeres in fungi, plants, mammals... and now fruit flies. "With the centromere sequences in hand, we are poised to leverage the powerful fruit fly genetic toolkit to understand the role these sequences play in centromere function and evolution," says Professor Barbara Mellone, co-lead author of the study.
More information: Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, et al. (2019) Islands of retroelements are major components of Drosophila centromeres. PLoS Biol 17(5): e3000241. doi.org/10.1371/journal.pbio.3000241
Journal information: PLoS Biology
Provided by Public Library of Science