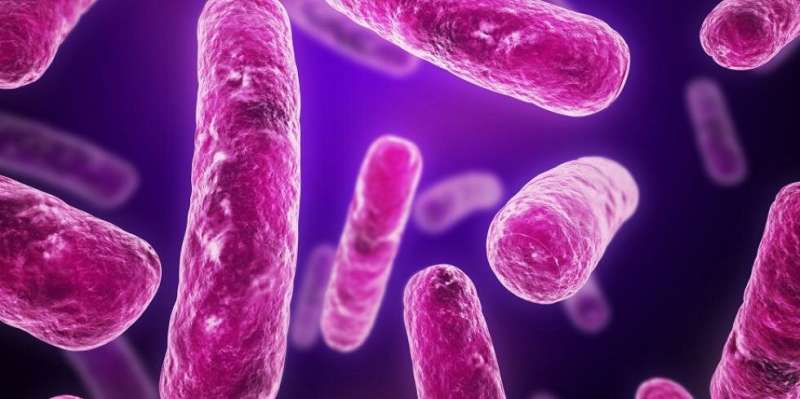
Visualisation of E. coli bacteria. Credit: University of Technology, Sydney
Urinary tract infections (UTIs) are among the world's most common bacterial infections, affecting around 50 per cent of women and five per cent of men. They can present as low-level cystitis or cause debilitating and potentially life-threatening conditions such as blood sepsis and kidney infection. Each year, treatment costs run to billions of dollars worldwide.
And they're becoming harder to treat, as the E. coli bacteria responsible for the infections become increasingly resistant to multiple antibiotics. According to new research from the Australian Centre for Genomic Epidemiological Microbiology (Ausgem), a partnership between the University of Technology Sydney (UTS) and the NSW Department of Primary Industries, the clue could well lie in the food we eat.
"Avian pathogenic E. coli (APEC) cause significicant economic losses to poultry production. There's also a growing body of evidence that some subsets of these avian pathogens have potential to cause disease in humans," explains Ausgem's UTS research lead, Professor Steven Djordjevic.
"APEC carry genetic elements called plasmids, which contribute to their ability to cause disease. Interestingly, these plasmids can move around between bacteria of the same species and between other gut-inhabiting bacteria and capture drug resistant genes as they go, with the potential to create new hybrid virulent strains."
The great plasmid migration
Last year, the Ausgem team made a novel discovery in bacterial isolates from a patient presenting with a UTI that later progressed to urosepsis – a form of sepsis where the source is localised to the urinary tract.
"The bacterial genome sequence information revealed a normal 'garden variety' commensal E. coli – not widely known to cause disease – that had acquired a plasmid that is related to plasmids normally associated with APEC. We concluded that the acquisition of this plasmid is likely to have played a role in converting this otherwise harmless E. coli into something more sinister."
While Djordjevic stresses it's difficult to establish absolute directionality in infection from chickens to humans, there is a growing weight of evidence to suggest these plasmids are moving around, finding their way into E. coli that inhabit our gastrointestinal tracts – and those of other food-producing animals. Even fresh produce such as salad items are susceptible through the soils and organic fertilisers they are grown in.
But it's not about changing what we eat; Ausgem's research shows that a multi-sector approach is crucial in understanding and addressing antibiotic resistance in our food chain. This means looking at how we use antibiotics in humans and in food-producing and companion animals, and how effluent is treated to remove multiple drug resistant bacteria and minimise or eliminate unmetabolised antibiotics that are excreted.
"Australia can be proud of its sound antimicrobial stewardship practices in agriculture," says Djordjevic. "But these can only go so far. Developing new vaccines and other intervention strategies to lower the overall use of antibiotics is still vital."
Professor Steven Djordjevic in the lab. Credit: Catherine McElhone
One Health approach to tackling resistance
With the world's population predicted to rise to about nine billion by 2050, the necessary boost in food production is predicted to drive greater reliance on antimicrobials. By the same year, antibiotic resistance is expected to account for around 10 million human deaths and between 60 to 100 trillion dollars in economic losses annually if nothing is done to curb antimicrobial drug resistance.
While the impetus in genomic sequencing in recent years has been on understanding pathogens that cause disease in humans, Ausgem are casting a much wider net – and revealing a new level of complexity and connection in tackling this critical global challenge.
"Antimicrobial resistance isn't just a problem in hospitals and aged care facilities," says Djordjevic. "It's inherent in how we produce food and treat effluent – safely and with minimal impact on the environment. We need to understand the role each part plays."
The team are looking at both how these bacteria colonise the gastrointestinal tract of our major food-producing animals, and how they intermingle with the environment and wildlife. Migratory bird species in particular play an increasingly important role in transmitting drug resistance genes across geographic borders.
This so-called One Health approach advocated by the World Health Organization recognises the intimate connection between the health of people, animals and the environment.
Advancing technologies
For Professor Aaron Darling, who is driving the computational and data generation aspects of the research, the project has offered an exciting opportunity to break new scientific ground.
"Advancements in high throughput DNA sequencing technologies have finally made it possible to effectively trace the movement and spread of bacteria in the environment," Darling explains.
"We've been developing innovative ways to generate and analyse the data, allowing us to carry out surveys at unprecedented scale and precision."
This research is but one arm of a connected and concerted effort by Ausgem to lead the charge in One Health surveillance of antimicrobial resistance and safeguard public health, biosecurity and food production. The centre is uniquely placed to leverage the exceptional knowledge, resources and skills of its academic and government partners in pursuit of a better future for people, plants, animals and the environment.
Provided by University of Technology, Sydney