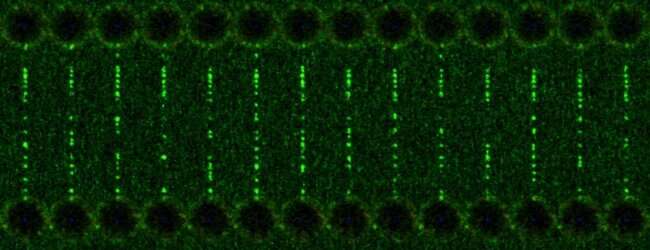
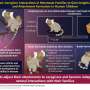
MRC LMS scientists show the off-target binding in a time-lapse. Credit: Prof David Rueda
Distortions to DNA, which occur routinely during gene expression and other cellular processes, could lead to off-target changes to the genome when using CRISPR-Cas9, a new study suggests. The Medical Research Council scientists behind the research say that their findings may help to pave the way to improve on the accuracy of gene editing for clinical applications.
During the expression of genes, DNA is stretched and distorted out of its usual shape. While this is needed for proper function of the cell's own machinery, it may pose a challenge for CRISPR-Cas9 gene editing by increasing the risk of off-target edits, potentially introducing harmful changes. The findings, from scientists at the MRC London Institute of Medical Sciences and AstraZeneca, are published today in the journal Nature Structural and Molecular Biology.
CRISPR-Cas9, a gene editing tool that allows researchers to find and edit strands of DNA, has gained worldwide recognition for its multitude of purposes as scientists use the technology across a range of sectors, including medicine, drug discovery and agriculture.
In the study, the accuracy and precision of CRISPR-Cas9 was investigated using a novel approach: scientists used optical tweezers – a tool that uses laser beams to manipulate DNA – to mimic the contortions that DNA naturally goes through as it's read by the cell's machinery. They then used CRISPR-Cas9 to edit the gene and monitored its accuracy using fluorescence microscopy.
Results showed that CRISPR is accurate when DNA is loose and relaxed. But when distortions occurred – in this case from being highly stretched – accuracy decreased, and off-target edits were observed. Understanding this effect will aid the design of CRISPR systems with increased accuracy, alongside methods to assess this risk.
Study lead author Professor David Rueda, of the MRC LMS and Imperial College London, said: "CRISPR-Cas9 has gained prominence as a potential tool to help prevent or combat diseases caused by mutations in DNA and we wanted to investigate how accurate it is. When we manipulated DNA structures, mimicking the natural contortions that DNA undergoes, we found edits were made to unintended sites. In this scenario, CRISPR-Cas9 can show as much as 50% mismatch in the targeting system which would be a concern for using this gene-editing tool in clinical settings.
"On the other hand, what's very positive is that we have developed a system that works to image CRISPR-Cas9 clearly so we can study why mistakes happen and adapt the technology to improve on accuracy and specificity."
Dr. Emanuela Cuomo, Associate Director, Discovery Sciences at AstraZeneca, said: "At AstraZeneca, we are working to understand the mechanism of action of CRISPR to enable us to design improved enzymes for research and potentially therapeutic purposes in the future. This research adds to our learnings of CRISPR-Cas9 and highlights future areas of exploration to better understand the relationship between DNA structure and off-target effects."
Dr. Lindsay Wilson, programme manager for genetics, epigenetics and genomics at the MRC, said: "This research is crucial because it helps us understand the risks of using CRISPR, for example to edit human cells. It might, ultimately, lead to strategies to reduce the risk, too. The MRC supports the use of scientifically and ethically rigorous laboratory studies using genome editing, including research to understand the technological risks and limitations, and steps to address these."
The scientists say their next steps are to determine how they may be able to reduce the chances for unintended edits. They plan to use their new system to further study CRISPR's accuracy and, ideally, develop off-target prediction algorithms based on their findings.
More information: Matthew D. Newton et al. DNA stretching induces Cas9 off-target activity, Nature Structural & Molecular Biology (2019). DOI: 10.1038/s41594-019-0188-z
Journal information: Nature Structural and Molecular Biology , Nature Structural & Molecular Biology
Provided by Medical Research Council