Nested CRISPR enables efficient genome editing using long DNA fragments
CRISPR is a technique that is revolutionizing biomedical research through high-precision genome editing. However, even though it allows the creation or correction of mutations consisting of a single or few nucleotides with relative ease, it still possesses limitations for larger fragments of DNA in the genome. For instance, the genomic insertion of a gene that produces a fluorescent protein such as the widely-used GFP suffers from poor efficiency and involves complicated cloning steps.
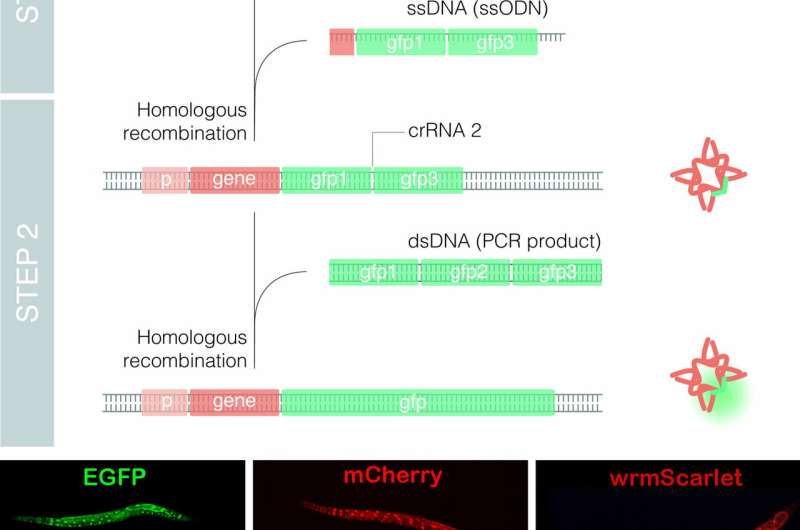
The group of Dr. Cerón at the Bellvitge Biomedical Research Institute (IDIBELL) used the model organism Caenorhabditis elegans to optimize the technique, leading to the development of a method called nested CRISPR. This cloning-free method involves the insertion of long DNA fragments in two steps. In the first step, a small portion (less than 200 nucleotides) of the long fragment is inserted in the genome. During the second step, this small fragment then serves as a "nest" or "landing pad" for the efficient insertion of the longer fragment (of approximately one kilobase).
This work, recently published in Genetics, a journal of the Genetics Society of America, is spawning unprecedented interest. Model organisms with a short life cycle such as C. elegans enable researchers to explore both the possibilities and limitations of CRISPR. Researcher Jeremy Vicencio, together with predoctoral researchers Carmen Martínez and Xènia Serrat, have performed hundreds of microinjections in the C. elegans germline and thousands of genotypings to solidly and convincingly demonstrate the efficiency of nested CRISPR.
Nested CRISPR utilizes commercial oligonucleotides for the first step of DNA repair and universal PCR products for the second. This facilitates large-scale experiments by making it possible to tag hundreds of genes with fluorescent proteins. Moreover, since nested CRISPR is modular, additional peptides or proteins of interest can also be integrated in combination with these fluorescent proteins. For example, the group is currently working on the tagging of these fluorescent proteins with peptides that allow their degradation in a controlled manner.
Finally, the group modeling diseases in C. elegans, directed by Dr. Cerón at IDIBELL, intends to use nested CRISPR to replace genes in C. elegans with their human counterparts. This would prompt the use of this small nematode as a multicellular model for studying the effect of human mutations that are associated with disease. This system would be of substantial interest in the field of personalized medicine since it could rapidly and efficiently provide a prognosis regarding the pathogenicity of a mutation or genomic variation (known as polymorphisms).
More information: Jeremy Vicencio et al, Efficient Generation of Endogenous Fluorescent Reporters by Nested CRISPR in Caenorhabditis elegans, Genetics (2019). DOI: 10.1534/genetics.119.301965
Journal information: Genetics
Provided by IDIBELL-Bellvitge Biomedical Research Institute