A new technique will make it possible to detect viruses in the field. In this case, a yam plot in Guadeloupe. Credit: D. Filloux, Cirad
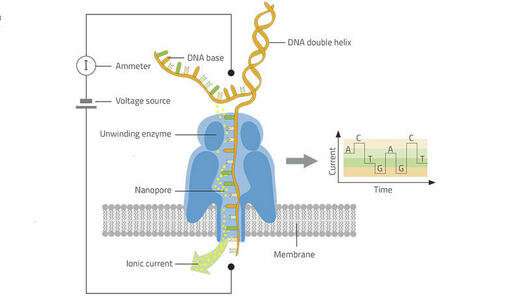
In a first for plant virology, a team from CIRAD recently used nanopore technology to sequence the entire genomes of two yam RNA viruses. This little-used but promising molecular biology technique paves the way for new tools for field diagnosis of plant, animal and human diseases.
A new, high-throughput, portable sequencing technique has been developed in recent years for human and animal health purposes. It uses mobile laboratories to diagnose viruses such as Ebola or Zika almost instantly, in the field. Diagnosis is quick and early, which avoids the need to transfer contaminated samples.
"The technology is characterized by the production of long nucleotide sequences, which makes it possible to sequence the entire viral genome," Philippe Roumagnac, a virologist with CIRAD, explains. CIRAD was one of the first laboratories in the world to test and validate its use in plant virology. "Using a diseased yam plant, it took us just a few hours to sequence the entire genome of two single-strand RNA viruses, a macluravirus and a potyvirus," his colleague Denis Filloux adds.
As with human virology, the fact that the technique has now been validated in a plant virology laboratory paves the way for real-time, mobile detection of chronic, seasonal or emerging plant viruses, even in isolated areas. By shortening the time that elapses between sampling and diagnosis, the technology will help epidemiosurveillance networks detect harmful organisms at an earlier stage.
Credit: Kerstin Göpfrich
More information: Denis Filloux et al, Nanopore-based detection and characterization of yam viruses, Scientific Reports (2018). DOI: 10.1038/s41598-018-36042-7
Journal information: Scientific Reports
Provided by CIRAD