Scientists find great diversity, novel molecules in microbiome of tree roots
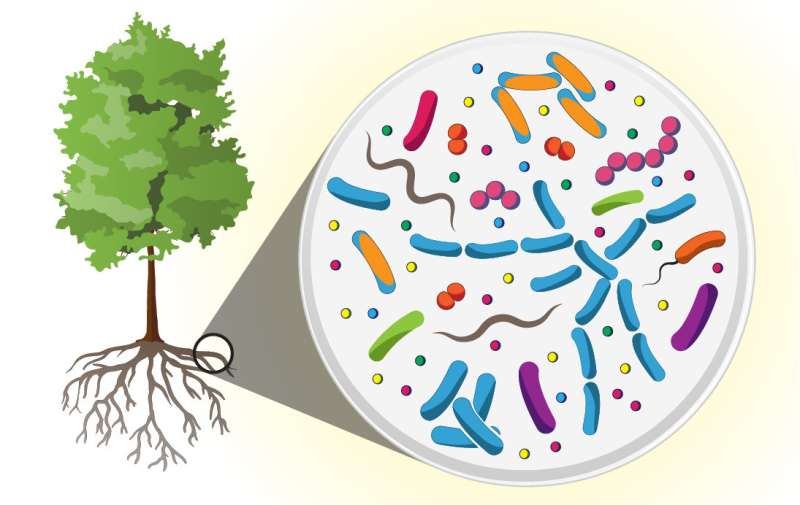
Researchers with the Department of Energy's Oak Ridge National Laboratory have discovered that communities of microbes living in and around poplar tree roots are ten times more diverse than the human microbiome and produce a cornucopia of novel molecules that could be useful as antibiotics, anti-cancer drugs, or for agricultural applications.
The study marks the first deep look at gene clusters in the Populus microbiome that code for the production of unique and varied natural products.
Microbes use natural products primarily as signals to communicate with other microbes or with the host plant. These molecules can trigger bacteria to grow, change, or slow their development. As competition for key resources can be fierce, microbes also secrete natural products as antibiotics to eliminate competitors.
The specific nature of these antibiotics, which often target select bacteria, makes them attractive candidates for use as targeted medicines that could kill the harmful species rather than a broad swath of microbes in a particular community, such as the human gut. The findings provide a starting point for exploring natural products and lay the groundwork for further investigation into the role these molecules play in plant-microbe interactions.
"We are focused on the exchange of energy, information, and materials across the plant-microbe interface," said Mitch Doktycz, lead for the Plant-Microbe Interfaces (PMI) Scientific Focus Area at ORNL. "Natural products are key to the performance and health of microbes and their host plants."
In surveying the breadth and diversity of natural products and the genetic underpinnings of the microbes producing them, the researchers found an abundance of gene clusters that do not fit into known groups. Almost 15 percent of the gene clusters do not fall into common structural categories. Of the clusters that do fit into known groups, only one percent match sequences that have been previously characterized.
Rooting out natural products
"That is a lot of biosynthetic potential," researcher Patricia Blair said. "Some of the unique gene sequences may code for known natural products. Some may code for molecules we've never seen before."
The research team conducted computational analysis of more than 300 bacterial genome sequences along with metagenomic samples from dozens of locations. This wealth of genomic information was gathered through previous ORNL research under PMI. The recent paper, published in mSystems, marks the first time many of these bacterial genomes have been described, although the sequences are regularly made available through the Joint Genome Institute.
At ORNL, these findings will guide ongoing research into the ways plants and microbes interact and influence each other's survival in the wild. In particular, researchers will be studying the effects of specific natural products and producers on constructed plant community dynamics.
"Some bacteria are just rife with these gene clusters," Blair said. "Scientists could prioritize a strain of bacteria or a class of natural products for further study based on these data. There is so much information contained in a single genome, that you just need a place to start."
More information: Patricia M. Blair et al. Exploration of the Biosynthetic Potential of the Populus Microbiome, mSystems (2018). DOI: 10.1128/mSystems.00045-18
Provided by Oak Ridge National Laboratory