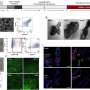
Fishing quotas upended by nuclear DNA analysis

For decades, mitochondrial DNA analysis has been the dominant method used to make decisions about fishing quotas, culling, hunting quotas, or translocating animals from one population of a threatened species to another.
A study published in Scientific Reports shows that while mitochondrial DNA analysis is very useful for a number of applications, analysing genetic differences for conservation purposes isn't one of them. The study highlights previously unrecognised genetic differences between populations of sardines.
In the study, researchers analysed mitochondrial DNA from southern hemisphere sardines caught off the western and southern coasts of South Africa.
"The mitochondrial DNA results indicated that overfishing the species in one part of its range would not create a major problem, as the population would be quickly replenished from elsewhere. But when we analysed far more sardine genes by sequencing a large portion of the nuclear genome, a completely different picture emerged," says Prof. Peter Teske from the Centre for Ecological Genomics and Wildlife Conservation at the University of Johannesburg.
For the study, the researchers analysed mitochondrial DNA and nuclear DNA for southern hemisphere sardines, a commercially exploited fish, and the Knysna sandgoby, a small endemic fish whose range is identical to that of the sardine.
Two kinds of DNA
Each living cell in an animal or plant contains two sources of DNA. Mitochondrial DNA (mtDNA) is easy to obtain from the mitochondria inside each cell. The mtDNA only has a few dozen genes, so it is quick and economical to sequence.
But nuclear DNA (nDNA) is far more difficult and expensive to work with. It only exists inside the nucleus of each cell. The nDNA can contain tens of thousands of genes, depending on the species. Luckily, sequencing costs today are fraction of what they were just a few years ago.
Plummeting costs
"At first, sequencing the whole human nuclear genome cost billions of U.S. dollars and took a consortium of researchers approximately 10 years to complete," says Arsalan Emami-Khoyi, a Postdoctoral Research Fellow at the Centre.
"Nowadays, it is possible to sequence the whole human nuclear genome within an hour, for less than a thousand dollars. So our centre contracts out nuclear DNA sequencing to commercial laboratories. And the gigabytes of data from the nDNA sequencing does not require investing in our own infrastructure—they were instead analysed at the state-run Centre for High Performance Computing in Cape Town," he adds.
"Taken together, it means we can now analyse much of the nuclear genome of our study organisms, which gives us much more robust results than was possible previously, when we sequenced only a handful of mitochondrial genes."
Previously unrecognised genetic differences
"Isolation by distance" is a concept that has been used for decades to decide indirectly whether the genetic makeup of populations of one species change with increasing geographic distance from each other. This indicates how well the different populations of a species are connected, which in turn influences fishing quotas and other conservation measures.
"This research indicates that a huge number of population genetic studies done in recent decades all over the world provided questionable results, because they were based on mitochondrial DNA," says Teske.
"Analysing mitochondrial DNA may indicate that there is no isolation by distance between the populations of a threatened or exploited species. When the populations are geographically close together, and the species is physically capable of covering the distance between the populations, that answer can look very convincing. This is what mitochondrial DNA told us about the southern hemisphere sardine," he adds.
Keeping fisheries open
But the present study shows that nuclear DNA analysis can highlight previously unrecognised genetic differences between the populations of a high-dispersal species, meaning a species that can cover geographic distances easily.
"As an example, our nuclear DNA analysis of the sardine showed that, contrary to what mtDNA is telling us, the level of isolation by distance is, in fact, very high. In practical terms, this means that if the west coast population becomes overfished, it will take a very long time for the south coast population to replenish it. The fishery could even be closed altogether," says Teske.
"For species that are threatened, or exploited, this means previous research on isolation by distance needs to be re-assessed for informed management decisions about quotas, culling and translocations. Past results need to be augmented with more sophisticated methods, such as partial sequencing of the nuclear genome," concludes Teske.
More information: Peter R. Teske et al, Mitochondrial DNA is unsuitable to test for isolation by distance, Scientific Reports (2018). DOI: 10.1038/s41598-018-25138-9
Journal information: Scientific Reports
Provided by University of Johannesburg