Autonomous gene expression control nanodevice will contribute to medical care
Gene expression is a fundamental of life, where each cell switches specific genes on and off. Thus, an autonomous device that could control the on-off switching would have great value in medical care.
Synthetic genetic circuits are a technology to control gene expression and program cells to perform desired functions. Therefore, increasing the complexity of the genetic circuit will allow us to control cell fates more accurately.
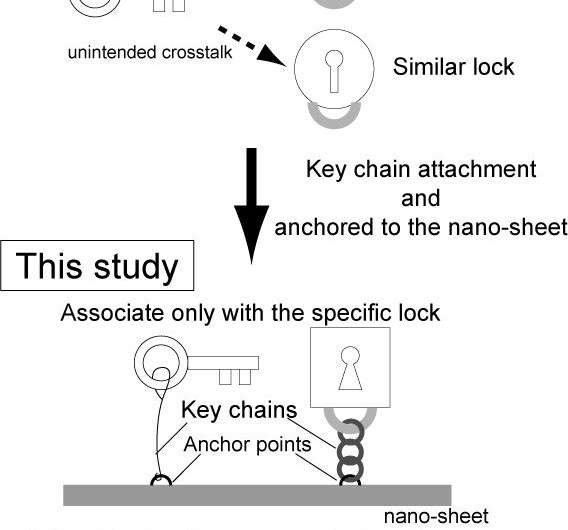
However, the complexity of genetic circuits remains low. This is because, in conventional reaction-diffusion systems, the enzymes and substrates are provided separately, and non-specific binding of the enzymes to the substrates causes unintended crosstalk among the different circuits.
Osaka University-led researchers, in a joint research project with The University of Tokyo, Kyoto University, and Waseda University, constructed integrated gene logic-chips called "gene nanochips." Using integrated factors on the nanochips, these self-contained nanochips can switch genes on and off within a single chip, preventing unintended crosstalk.
The researchers showed the autonomous responses of the nanochips in artificial cells: environmental sensing, information computation and product output at the single-chip level. Their research results were published in Nature Nanotechnology.
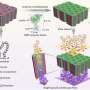
DNA nanotechnology is a versatile method used to construct custom structures and to control precise molecular layouts. The researchers used a rectangular sheet (90 nm wide, 60 nm deep, 2 nm high), and integrated enzyme, RNA polymerase (RNAP, an enzyme that synthesizes RNA from a DNA template), and multiple target-gene substrates.
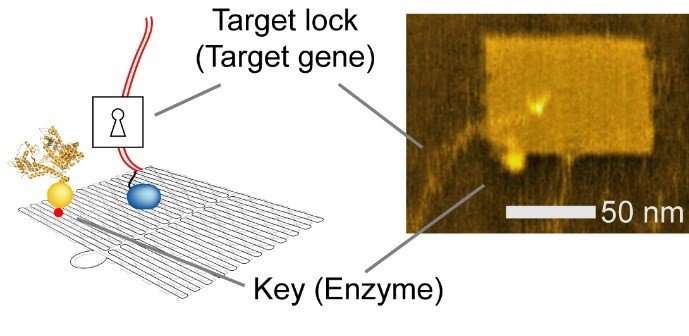
The nano-layout ability of DNA nanotechnology allows the researcher to rationally design gene expression levels by changing the intermolecular distances between the enzyme and the target genes, thus affecting the collision efficiency and subsequent reaction.
The researchers further integrated sensors. Ideally, a sensor that is capable of detecting any type of signal should have minimal design limitations. However, conventional methods have suffered from several limitations (e.g., materials). This is because, in conventional genetic circuits, the sensor is part of the substrate of the enzyme (e.g., DNA in transcription; see Note for detail).
Conversely, in this study, the sensor part was independent of the enzymatic reaction. Thus, the researchers can use any sensor materials that change the effective intermolecular distance on signal recognition, allowing the construction of various sensors responding to distinct signals (microRNAs, chemical compounds, proteins and light). Moreover, by combining and integrating sensors responding to distinct signals, the researchers have succeeded in photo-reprogramming of the genetic circuits.
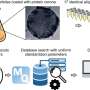
Finally, the nanochip allowed the researchers to simplify the construction of a genetic circuit that responded to an artificial cell, a water-in-oil droplet, and they could compute its miRNA profile, by simply mixing the orthogonal chips, expanding the power of the genetic circuit.
Corresponding author Hisashi Tadakuma says, "All factors necessary for transcription reactions are on this integrated nanochip, so environmental sensing, information computation, and product output can be completed at the single-chip level. In the near future, autonomous nanochips will be useful in maintaining the cell in the healthy state through controlling gene expression spatially and temporally, which will embody the ideal of the saying 'prevention is best cure'. "
Ideally, a sensor that is capable of detecting any type of signal should have minimal design limitations. However, conventional methods have suffered from several limitations (e.g., materials). This is because, in conventional genetic circuits, the efficiency of binding between an enzyme such as RNAP (key) and a target gene (target lock), specifically a promoter sequence on the DNA molecule, is controlled by transcription factors (regulators). As regulators and enzymes should bind to the sensor (operator region) near the target lock, the material of the sensor has to be a substrate of the regulators and enzymes (e.g., DNA in transcription), and the regulator should be a DNA-binding protein. Thus, both the regulator (signal) and sensor have material limitations.
However, in this study, the binding frequency and subsequent reaction are controlled by (1) the intermolecular distance between an enzyme and a target gene, and (2) the characteristics of the key chain (e.g., rigidity, length). Thus the sensor part is independent of the enzymatic reaction, and the sensor and regulators have no material limitations, allowing the various sensors responding to distinct signals (chemical compounds, proteins and light).
More information: Takeya Masubuchi et al. Construction of integrated gene logic-chip, Nature Nanotechnology (2018). DOI: 10.1038/s41565-018-0202-3
Journal information: Nature Nanotechnology
Provided by Osaka University