Hi-res image of structure of the origin recognition complex bound to DNA revealed
Cells propagate by making copies of themselves via genome replication. Arguably, replication of DNA is the most fundamental and conserved mechanism of all life forms. Cracking the secret of how this process is achieved with the utmost accuracy is the key to understanding the secret of life. When Watson and Crick first proposed how DNA is replicated based on the structure of the DNA double helix more than a half-century ago, many believed that the structure of the machinery that separates the two strands of DNA for replication would be forthcoming. However, the replication machine turns out to be much more complicated than previously imagined because of its large size, its tripartite nature (it is made up of three engines) and flexibility. Structural information for the DNA replication machinery at atomic resolution by conventional methods was not available until very recently with the advent of the resolution revolution of cryo-EM technology.
A series of articles published by the Tye (HKUST)/Gao (Peking University) collaboration opens the door for deciphering the function of the DNA replication machinery at unprecedented resolutions. The first, published in Nature 2015, determined the structure of the core engine of the DNA replication machine called the MCM complex. The second reported an open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. The third now appears in Nature, detailing the atomic structure of the Origin Recognition Complex (ORC) that selects start sites genome-wide to initiate DNA replication.
Every human came from a single cell (fertilized egg) after approximately 1016 cell divisions. Each cell division requires the exact replication of the genome such that each daughter cell receives a complete complement of the identical genetic information in the form of DNA. Aberrant DNA replication that results in deregulated cell divisions is the cause of many cancers and developmental disorders. Genome replication is equally important in the maintenance of living organisms as all cells have "expiration dates" and most are replenished by stem cells that retain the capability to divide.
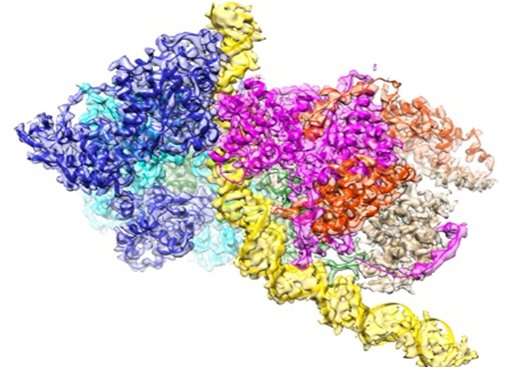
Aging is also a general phenomenon of the breakdown of the replication machinery, either in repair synthesis of damaged DNA or in the fidelity of whole genome replication. In a study led by Professor Bik Tye and Dr. Yuanliang Zhai at HKUST, with Professor Ning Gao at Peking University, the structure of the machine called Origin Recognition Complex (ORC) that initiates DNA replication was determined at atomic resolution for the first time using cryo-electron microscopy. This structure explains how ORC is able to scan a sea of bases (DNA is made up of 4 bases, A, T, G, C) to select the correct sites programmed for DNA replication to begin. It is believed that indiscriminate selection of too many sites may lead to rapid replication of the genome and therefore rapid cell divisions, a characteristic of cancer cells. In contrast, inefficient selection of sites resulting in sluggish cell divisions especially at critical junctures of human development may lead to developmental disorders.
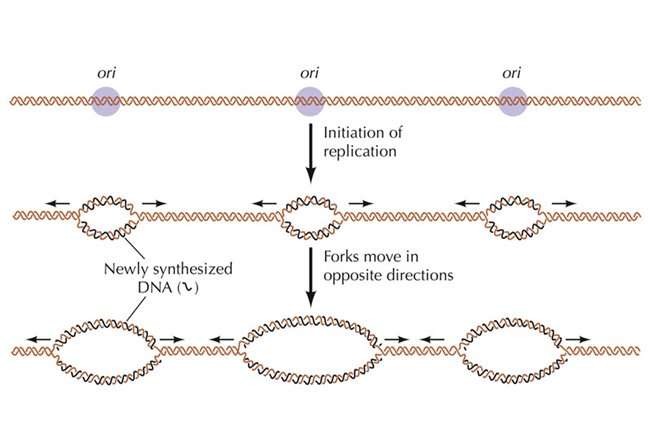
A case in point is the Meier-Gorlin Syndrome (MGS), a rare form of hereditary dwarfism that is characterized by prenatal stunted growth and postnatal proportionate short stature. Interestingly, mutations associated with the MGS are located in five genes (ORC1, ORC4, ORC6, CDT1 and CDC6), all of which are components of the DNA replication initiation machine. Afflicted individuals with ORC1 and ORC4 mutations appear to have the most severe short stature. In the study that appears in the current issue of Nature, Tye/Gao showed that of the six subunits that form the ORC complex of the replication initiation machine, ORC1 and ORC4 play the determinant role in the selection mechanism for initiation sites.
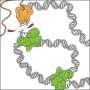
Fundamentally, the most important function of ORC is to recruit the MCM double hexameric complex, the catalytic core of the DNA helicase that separates duplex DNA, onto origin DNA. The atomic structure of ORC bound to DNA reported in this issue of Nature reveals that bending of DNA by ORC provides a docking surface for the insertion of DNA into the open ring of the MCM helicase. Furthermore, it reveals that origin DNA is selected for its unique structure rather than specific base sequence. These novel findings help explain how ORC selects replication origins at unique sites in the genome that are not predictable solely by their base sequences.
DNA replication is a defining feature for all living organisms and the machine that carries out this function is conserved from fungus to plants to human. Understanding the atomic structure of the DNA replication machine (or any bio-molecular machines) is fundamentally important because all applied technology and engineering are founded in basic science/knowledge. For example, a three dimensional view of the DNA replication machine at 3Å resolution may help us identify better targets for cancer therapy such that synthetic chemicals can be custom made to fit the target. More importantly, structures help us to fully understand the mechanistic functions of molecular machines and therefore the roots of diseases due to suboptimal functions of these machines. Toward this goal, the Hong Kong University of Science & Technology will be establishing a state of the art cryo-electron microscopy facility for the study of high resolution structures of bio-molecular machines.
More information: Ningning Li et al, Structure of the origin recognition complex bound to DNA replication origin, Nature (2018). DOI: 10.1038/s41586-018-0293-x
Ningning Li et al. Structure of the eukaryotic MCM complex at 3.8 Å, Nature (2015). DOI: 10.1038/nature14685
Yuanliang Zhai et al. Open-ringed structure of the Cdt1–Mcm2–7 complex as a precursor of the MCM double hexamer, Nature Structural & Molecular Biology (2017). DOI: 10.1038/nsmb.3374
Matthew L. Bochman et al. Strand separation unravelled, Nature (2015). DOI: 10.1038/nature14643
Journal information: Nature , Nature Structural & Molecular Biology
Provided by Hong Kong University of Science and Technology