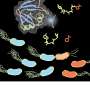
Researchers discover mechanism behind citrus canker bacteria's defense system for predators
Xanthomonas citri, the bacterium that causes citrus canker, a disease responsible for major damage to lemon and orange groves in worldwide scale, has a veritable arsenal of weapons to overcome constant competition with other bacterial species and ward off natural predators such as amoebae.
Despite the fact that citrus canker have already caused significant economic losses, X. citri's ability to persist in the environment is still poorly understood. However, a new study performed in Brazil perfects scientific understanding on the gene expression regulating the bacteria's defense mechanism against natural predators—which might subsidize the development of strategies to fight the pest on infected plants.
Researchers with the State University of Campinas's Biology Institute (IB-UNICAMP) and the University of São Paulo's Chemistry Institute (IQ-USP) have demonstrated a previously unknown mechanism used by X. citri against amoebae, the main source of selective pressure on bacteria in the natural environment.
The study revealed that the mechanism resides in a protein secretion system known as the T6SS (type VI secretion system) present in several species of bacterium but only now characterized in X. citri. It is, basically, a complex of proteins injected into the amoeba through the bacterial envelope.
"X. citri has several secretion systems. The one we studied is a kind of contractile machine that projects out through the bacterial cell membrane and secretes toxins or proteins into the target cell, in this case, the amoeba," said Cristina Alvarez-Martinez, a researcher affiliated with IB-UNICAMP and co-author of an article published in the journal Environmental Microbiology.
Signaling pathway
In addition to demonstrating that the mechanism is used for resistance to the amoeba Dictyostelium discoideum, the researchers also discovered a new signaling pathway that controls the expression of the secretion system's genes in response to contact with the amoeba. Although the signaling pathway is also found in the genomes of other environmental bacteria, this is the first time it has been studied in X. citri.
"The paper describes a new mechanism for regulating gene expression also used by other bacteria in the natural environment," Alvarez-Martinez said.
The researchers found that the translocation of proteins occurs in a manner controlled by the bacterium. "In the paper, we demonstrate that the bacterium induces transcription of T6SS genes to produce the secretion channel using this new signaling mechanism we identified. The signaling mechanism is activated in response to contact with the amoeba," Alvarez-Martinez said.
Genetically-induced vulnerability
The bacterium's genome was sequenced in 2001 via FAPESP's Genome Program, later enabling localization of the genes that encode the T6SS secretion mechanism. As a result, the researchers were able to modify the bacterium genetically so that it stopped producing T6SS.
The study compares two strains of the bacterium: a mutant strain without the T6SS genes and a wild-type strain with no genetic modification. The conclusion was that the amoeba D. discoideum feeds more efficiently on the wild-type strain.
"When these genes were removed, the bacterium stopped producing the secretion system," Alvarez-Martinez explained. "We saw from the comparison that the wild-type strain displayed far greater resistance to the amoeba and survived better than our mutant strain without the secretion system. We succeeded in demonstrating that this is crucially important to the survival of X. citri."
Arsenal
According to Alvarez-Martinez, the study confirms that X. citri has an arsenal of mechanisms categorized in terms of secretion system families to withstand the attacks of environmental competitors.
"Many of these systems have already been studied. The bacterium's T3SS or type III secretion system, for example, is directly linked to its virulence. If this system is eliminated, it can't cause citrus canker. Its T4SS is involved in the task of eliminating other species of bacterium that compete with it for space and nutrients. Its T6SS, which we're now studying, acts as a resistance mechanism in disputes with predators, in this case, amoebae," said the FAPESP-supported researcher.
More research is necessary to completely understand what happens in this dispute between X. citri and D. discoideum. "One of the hypotheses raised in our paper is that the amoeba may be a sort of reservoir for bacteria. X. citri can kill the amoeba or multiply inside it. What we want to find out now is how the relationship between these two microorganisms is established," she said.
Whether or not this hypothesis is confirmed, one thing is certain: the recently identified secretion system increases the bacterium's resistance. This could have a strong impact on its persistence in the soil and on the leaves of citrus plants.
"The more we know about the mechanisms used by the bacterium to withstand adversity, the easier it will be to design ways and means of preventing the spread of citrus canker. In the future, our understanding of T6SS can help us think about a form of intervention to stop citrus canker from developing in the field, as well as contributing significantly to our understanding of X. citri's biology," Alvarez-Martinez said.
More information: Ethel Bayer-Santos et al, Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF σ factor and cognate Ser/Thr kinase, Environmental Microbiology (2018). DOI: 10.1111/1462-2920.14085
Journal information: Environmental Microbiology
Provided by FAPESP