Making protein patterns robust

Correct protein localization is crucial for many fundamental cellular processes. LMU physicists have now asked how to confer robustness against variations in protein concentrations on pattern formation mechanisms.
Many of the fundamental processes observed in cells depend on proper localization of proteins. For example, the division plane at which cell division takes place is marked by correct patterning of specific proteins, and should be robust against alterations in the relative concentrations of the proteins involved. Ludwig Maximilian University physicists led by Professor Erwin Frey, in collaboration with the team of Professor Petra Schwille (Max Planck Institute for Biochemistry, Munich) have now explored how this kind of stability is achieved. Their findings appear in the journal PNAS.
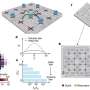
One of the most popular model systems for the study of biological pattern formation is the Min system, which restricts the plane of cell division (defined by a ring of the protein FtsZ) to mid-cell in in the rod-shaped bacterium Escherichia coli. The system comprises a small set of Min proteins whose distributions dynamically oscillate between the poles of the cell. Their net effect is to prevent assembly of the division ring on the membrane near the poles, thus confining FtsZ to the middle of the cell. The Min oscillations are set up by a complex interplay between the ATPase enzyme MinD and its activator MinE. MinE activates the enzymatic activity of membrane-bound MinD, thus triggering the conversion of its bound nucleotide adenosine triphosphate (ATP) to adenosine diphosphate (ADP), which causes the release of MinD-ADP into the cell cytoplasm. MinD then diffuses in the cytoplasm until ADP is replaced by ATP, which allows it to bind to the cell membrane once again. "For conventional mathematical models of this process pattern formation of MinE and MinD on the membrane can only work if the concentration of MinE is less than that of MinD," says Jonas Denk, a Ph.D. student in Frey's team and joint first author of the new paper. However, E. coli cells normally contain approximately equal levels of MinE and MinD, and in vitro experiments have shown that patterns can emerge even when the concentration of MinE is significantly increased.
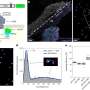
Denk and his coauthors have resolved this conflict between theory and experiment by incorporating into their model the effects of a conformational change in MinE, which occurs when the activator binds to the membrane. It has recently been shown that MinE occurs in two different structural forms, an open ('reactive') and a closed ('latent') conformation. The closed form has a low affinity for MinD, i.e. the probability that it binds to MinD is quite small. However, when it does bind, MinE is rapidly converted into the open form, which has a high affinity for MinD-ATP. Once in this state, MinE activates MinD's ATPase function, and both MinE and MinD-ADP are released from the membrane. The researchers suggest that the active form of MinE briefly remains in the open conformation and can therefore "jump on" the nearest membrane-bound MinD. If it cannot find one fast enough, it reverts to the closed low-affinity conformation. "This ability to switch between the two conformations is essential for the robustness of pattern formation, and allows pattern formation even for strongly enhanced MinE concentrations, as our simulations have shown," says Frey. Indeed, in vitro experiments carried out by Simon Kretschmer (a member of Schwille's group) confirmed the results of these simulations.
This modified model yields new insights into the mechanisms that regulate biological pattern formation, which have implications for other pattern-forming systems. Frey and colleagues argue that robustness in the face of changes in the concentrations of components could provide an evolutionary advantage, insofar as it endows cells with a higher degree of flexibility. Extending the tolerable margin compatible with pattern formation of error ensures that patterns can still be formed when relative protein levels are altered.
More information: MinE conformational switching confers robustness on self-organized Min protein patterns. PNAS. DOI: 10.1073/pnas.1719801115
Journal information: Proceedings of the National Academy of Sciences
Provided by Ludwig Maximilian University of Munich