March 29, 2018 report
ScarTrace helps understand how multicellular organisms develop from embryonic progenitors

A team of researchers with Oncode Institute, Hubrecht Institute-KNAW and University Medical Center Utrecht in The Netherlands has developed a new method to conduct whole-organism clone tracing using single-cell sequencing. In their paper published in the journal Nature, the group describes how they used their new method to conduct research on barcoded zebrafish cells.
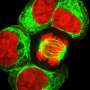
As the researchers note, embryonic development is an important stage for highly complicated organisms such as humans—only a very limited number of embryonic progenitors somehow manage to produce all of the cells that wind up in the adult body. To understand how this process works, the researchers further note, methods are required to measure the clonal history that occurs, and at the same time, perform cell identification at single-cell resolution. In this new effort, the researchers developed such a technique called ScarTrace. The name comes from a part of the technique that involves adding tandem copies of a fluorescent protein transgene that makes it possible to identify "scars" left behind in a transcription caused by CRISPR-Cas9 gene editing.
Using the technique, the researchers were able to track the clonal roots and cell outcomes of coded zebrafish cells. More specifically, they were able to trace back adult cells from several sites such as the kidneys, eyes and fins to specific progenitors. They note that the technique was sensitive enough to study the process that occurs when a progenitor commits to producing a left or right eye. They also found that cells in the skin and the caudal fin arose from the same progenitors. They report the identification of immune cells in fish fins with a distinct clonal origin from other kinds of blood cells.
The researchers suggest methods like theirs will help with the ultimate goal of tracing all the events that lead from a single cell to a fully developed body. Bolstering that claim was work by another team with members from China, the U.K. and the U.S.—they published a paper in the same Nature issue detailing their own work with a single-cell sequencing method they had developed called MAP-seq.
More information: Anna Alemany et al. Whole-organism clone tracing using single-cell sequencing, Nature (2018). DOI: 10.1038/nature25969
Abstract
Embryonic development is a crucial period in the life of a multicellular organism, during which limited sets of embryonic progenitors produce all cells in the adult body. Determining which fate these progenitors acquire in adult tissues requires the simultaneous measurement of clonal history and cell identity at single-cell resolution, which has been a major challenge. Clonal history has traditionally been investigated by microscopically tracking cells during development, monitoring the heritable expression of genetically encoded fluorescent proteins and, more recently, using next-generation sequencing technologies that exploit somatic mutations4, microsatellite instability, transposon tagging, viral barcoding, CRISPR–Cas9 genome editing and Cre–loxP recombination. Single-cell transcriptomics provides a powerful platform for unbiased cell-type classification. Here we present ScarTrace, a single-cell sequencing strategy that enables the simultaneous quantification of clonal history and cell type for thousands of cells obtained from different organs of the adult zebrafish. Using ScarTrace, we show that a small set of multipotent embryonic progenitors generate all haematopoietic cells in the kidney marrow, and that many progenitors produce specific cell types in the eyes and brain. In addition, we study when embryonic progenitors commit to the left or right eye. ScarTrace reveals that epidermal and mesenchymal cells in the caudal fin arise from the same progenitors, and that osteoblast-restricted precursors can produce mesenchymal cells during regeneration. Furthermore, we identify resident immune cells in the fin with a distinct clonal origin from other blood cell types. We envision that similar approaches will have major applications in other experimental systems, in which the matching of embryonic clonal origin to adult cell type will ultimately allow reconstruction of how the adult body is built from a single cell.
Journal information: Nature
© 2018 Phys.org