Credit: CC0 Public Domain
A team of researchers at Rockefeller University has discovered a new family of antibiotics by conducting a genetic study of a wide range of soil microorganism antibiotics. In their paper published in the journal Nature Microbiology, the group describes their study and how well samples of the new antibiotic worked in rats.
Bacteria are evolving to become drug resistant, making it increasingly difficult to treat people with bacterial infections. Because of this, scientists the world over are looking for new antibiotics. In this new effort, the researchers studied microorganisms that live in soil as a possible source of new antibiotics—in order to survive, they, too, must have some means of fighting off bacterial infections.
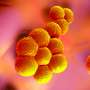
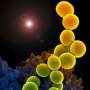
The researchers collected soil samples, then picked an antibiotic, daptomycin, to serve as a form of guide. It uses calcium to disrupt bacterial cell walls, killing the cells. The team then used DNA information that encodes for production of the antibiotic in daptomycin as a guide as they studied the genomes of antibiotics used by microorganisms in over 2000 soil samples. They came across a new family of antibiotics, one they named malacidins—they fight off infections by using calcium to disrupt bacterial cell walls. The team notes that unlike other antibacterial agents, the calcium does not cause the walls to leak, but instead disrupts their behavior in other ways.
To see how well the newly discovered antibiotics might work, the team tested some samples on rats that with induced MRSA skin infections. They report that the antibiotics completely eliminated the bacteria. The team also put the antibiotics through testing to determine whether bacteria could become resistant to malacidins—they report that they were unable to induce resistance in the lab thus far.
Much more testing with the new family of antibiotics is required before clinical trials, of course, but for now, the researchers are optimistic, suggesting many more members of the family are likely to be found.
More information: Bradley M. Hover et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens, Nature Microbiology (2018). DOI: 10.1038/s41564-018-0110-1
Abstract
Despite the wide availability of antibiotics, infectious diseases remain a leading cause of death worldwide1. In the absence of new therapies, mortality rates due to untreatable infections are predicted to rise more than tenfold by 2050. Natural products (NPs) made by cultured bacteria have been a major source of clinically useful antibiotics. In spite of decades of productivity, the use of bacteria in the search for new antibiotics was largely abandoned due to high rediscovery rates2,3. As only a fraction of bacterial diversity is regularly cultivated in the laboratory and just a fraction of the chemistries encoded by cultured bacteria are detected in fermentation experiments, most bacterial NPs remain hidden in the global microbiome. In an effort to access these hidden NPs, we have developed a culture-independent NP discovery platform that involves sequencing, bioinformatic analysis and heterologous expression of biosynthetic gene clusters captured on DNA extracted from environmental samples. Here, we describe the application of this platform to the discovery of the malacidins, a distinctive class of antibiotics that are commonly encoded in soil microbiomes but have never been reported in culture-based NP discovery efforts. The malacidins are active against multidrug-resistant pathogens, sterilize methicillin-resistant Staphylococcus aureus skin infections in an animal wound model and did not select for resistance under our laboratory conditions.
Journal information: Nature Microbiology
© 2018 Medical Xpress