Detecting new proteins in active brains of mice
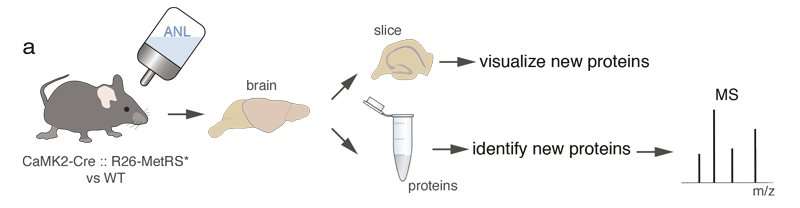
The complexity of living things is driven, in large part, by the huge diversity of cell types. Since all cells of an organism share the same genes, the diversity of cells must come from the particular proteins that are expressed. Cells in the brain are generally divided into neurons and glia. Within these two categories, however, lies a large diversity of cell types that we are only beginning to discover. The diversity of cell types in brain and other tissues has recently been expanded by new techniques, like RNA-sequencing, that identify and measure the mRNAs present in a cell, the so-called transcriptome. Although mRNAs are the template for proteins, the transcriptome is a poor proxy for proteins that a cell actually makes, the proteome. Scientists from the Max Planck Institute for Brain Research in Frankfurt now developed new methods to detect real-time changes in the proteome.
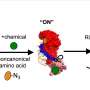
Building on prior technology, developed by the Lab of Erin Schuman at the Max Planck Institute and collaborators David Tirrell from Caltech and Daniela Dieterich from Magdeburg University, Beatriz Alvarez-Castelao and colleagues took advantage of a protein "metabolic" labeling system. In this system proteins during synthesis are "tagged" with an amino acid, which is, under normal conditions, not present in these cells. In order to label proteins in a particular cell type exclusively, the research team used a mutant methionyl tRNA synthetase (MetRS) that recognizes the modified amino acid. They then created a mouse line in which the MetRS can be expressed in specific cell types. When the non-canonical amino acid is administered to the mutant MetRS mice via the drinking water, only proteins in cells expressing the mutant metRS are labeled.
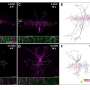
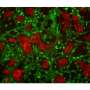
The proteins labeled in cells can be visualized and recognized with antibodies or can be extracted and identified using mass spectrometry. Alvarez-Castelao: "We used the technique to identify two different sets of brain proteins, those present in excitatory neurons in the hippocampus, a brain structure important for animal navigation and learning and memory, and inhibitory neurons in the cerebellum, a structure involved in motor behavior."
A particularly striking feature of this technology is that one can detect directly changes in brain proteins in response to a modified environment. Mice that were raised in an enriched sensory environment with a labyrinth, running wheel, and toys of varied textures showed significant changes in the proteome in the hippocampus, particularly in proteins that work at neuronal synapses. Schuman: "We think that, by combining this mouse with other "disease" mouse models, this method can be used to discover the proteins in particular cell-types and how proteomes change during brain development, learning, memory and disease."
More information: Beatriz Alvarez-Castelao et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo, Nature Biotechnology (2017). DOI: 10.1038/nbt.4016
Journal information: Nature Biotechnology
Provided by Max Planck Society