Researchers describe mechanism behind flagellar motility
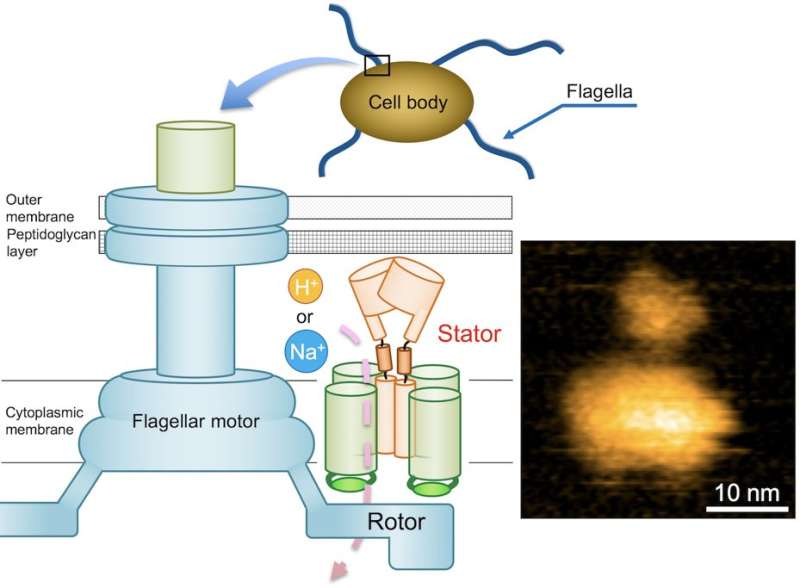
Bacteria swim in many different ways, and the motors that drive their swimming are widely varied, implying an adaptive response to an environment. One of the most commonly identified of such motors is flagella. Although providing motility is a primary feature of flagella, adhesive and other properties have also been attributed to them.
The flagellar motor consists of a rotor and a dozen stator units, and is driven by the energy created by ions when they migrate across the cell membrane. While the flagellar motor of some freshwater and enteric bacteria, such as Salmonella, is powered by protons (hydrogen ions, H+), which consists of four copies of the protein MotA and two copies of MotB, motors of other bacteria that live in salt water environments are powered by sodium ions (Na+).
In the case of Bacillus subtilis (B. subtilis) , a bacterium commonly found in the soil, the flagellar motor has two distinct types of stator units: H+-type MotAB and Na+-type MotPS. While the components are identified, the mechanism that powers the flagellar motor remains unknown. This mystery is further compounded by the fact that flagella are not clearly visible under the light microscope. These challenges set a team of Osaka University-centered researchers on a quest to elucidate the unknown mechanism. They reported their findings in Science Advances.
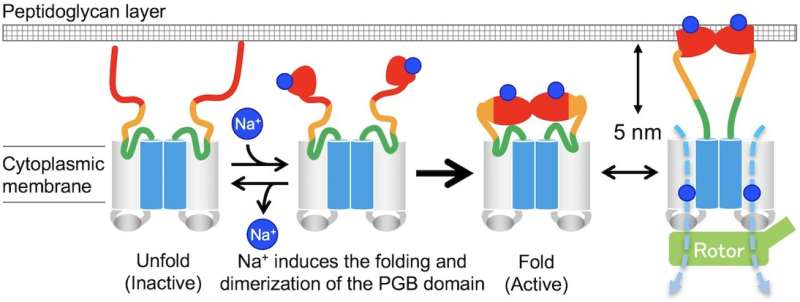
"Precise analyses of structural dynamics of the flagellar motor using high-speed atomic force microscopy (HS-AFM) combined with mutational analysis provided insights into the torque generation mechanism of the flagellar motor," the first author of the study Naoya Terahara says. "We made real-time observations of the Na+-induced structural changes to the domain of MotS that binds peptidoglycan, a polymer consisting of sugars and amino acids in bacteria."
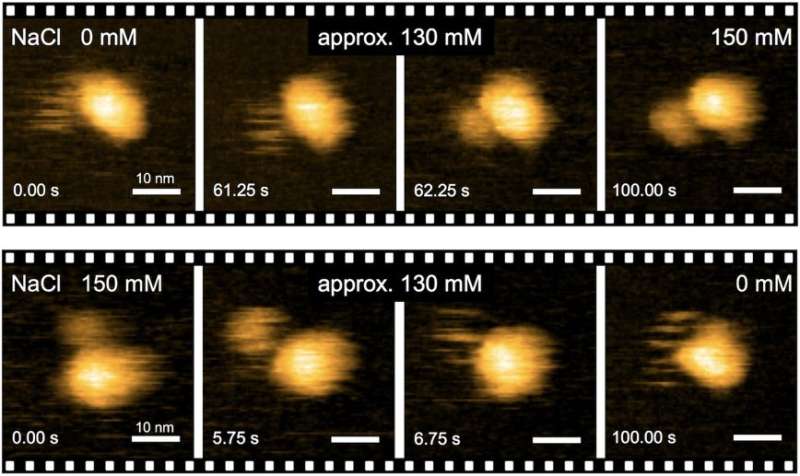
To understand how the MotPS protein complex responds to changes in the environment, the researchers first characterized the torque-speed relationship of the B. subtilis motor over a wide range of Na+ concentrations. The maximum speed of the motor was decreased from about 200 to 80 Hz when the external Na+ concentration was increased from 0 to 200 mM, although the stall torque was not changed at all.
Notably, when the domain of MotB that binds to peptidoglycan was replaced by that of MotS, the chimeric motor maintained the same torque-speed curve as the MotAB motor in the presence of Na+ ions but not in the absence. This means that Na+ is the main driver of the assembly and activation of flagellar motor, regardless of its composition.
"In the absence of sodium ions, MotPS exists as an inactive form in the cytoplasmic membrane because MotSc, the C-terminal periplasmic domain of MotS, adopts an unfolded conformation. When the concentration of sodium ions is increased, the binding of the ions to MotSc induces the folding of the PGB domain and facilitates its dimerization in a highly cooperative manner," corresponding author Tohru Minamino says.
Through this study, the team has also demonstrated the feasibility of using HS-AFM combined with mutational analysis to study intricate bacterial structures, which can provide valuable insights for industrial production of commercially viable products, such as proteases and therapeutics, as in the case of the B. subtilis.
More information: Naoya Terahara et al. Na + -induced structural transition of MotPS for stator assembly of the Bacillus flagellar motor, Science Advances (2017). DOI: 10.1126/sciadv.aao4119
Journal information: Science Advances
Provided by Osaka University