Artificial enzyme functions as well as natural version
Scientists from the University of Bristol have designed an artificial enzyme that functions as well as (and in some cases better than) a vital class of natural enzymes.
Enzymes are fundamentally important biological molecules that perform the bulk of the chemical reactions in all living organisms.
These reactions power cellular life and are involved in a great number of processes necessary to give cells their chemical and physical characteristics.
Furthermore, many enzymes participate in chemical reactions which have commercial or medical value, and for which we have no good manmade substances that can catalyse such reactions with the same precision and efficiency as enzymes.
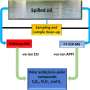
The artificial enzyme was synthesised in a fully functioning form by E. coli bacteria and could be of significant interest to the biotech industry due to its industrially-relevant catalytic activities, thermal stability and chemical resistance.
They could even be used as replacements for certain natural enzymes that are more complex and difficult to produce on a larger scale.
To achieve this the team used an approach that eliminates evolutionary complexity by employing small, simple and completely artificial proteins into the design process.
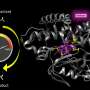
Dr Ross Anderson from the University of Bristol's School of Biochemistry, said: "We describe these proteins as maquettes, a term used in sculpture to denote a scale model from which a final work is created.
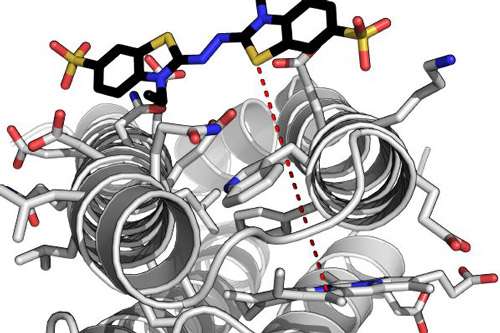
"The protein maquette similarly allows the designer to incorporate functional elements onto a stable protein scaffold. In this case, we adapted an earlier maquette design where we used the machinery present in a living bacterial cell to permanently graft the heme molecule into the protein's structure."
The alterations made, facilitated the binding of hydrogen peroxide to the heme molecule, a necessary step in catalysis common to the peroxidase family of enzymes.
With these changes, the maquette was capable not only of binding hydrogen peroxide, but also of converting it into a more reactive form that can pull electrons from a range of simple organic molecules, the same way that natural peroxidases function.
Because the maquette is also fully assembled in bacterial cells, it provided the team with the opportunity to exploit these chemical transformations in living cells, or to consider integrating it into the metabolic pathways of an organism.
Dr Anderson added: "So in essence, we have made an artificial (or de novo) enzyme that functions as well as (and in some cases better than) a vital class of natural enzymes.
"It is promiscuous - as many of the natural peroxidases are - and could stand as a robust prototype from which many functional catalysts could be developed.
"In catalytic activity, it is so close to natural enzymes (and given the fact that the bacteria make it in an active form), if it were purified without knowledge of its artificial origins it would be ascribed a natural function and treated as a novel natural enzyme."
More information: Daniel W. Watkins et al. Construction and in vivo assembly of a catalytically proficient and hyperthermostable de novo enzyme, Nature Communications (2017). DOI: 10.1038/s41467-017-00541-4
Journal information: Nature Communications
Provided by University of Bristol