'Twinkling' enzymes could light the way to better cancer drugs
A new test to show the properties of biologically important enzymes could help to streamline development of new treatments.
"Twinkle, twinkle, little kinase. How I wonder what form you are…"
It may not make for the best nursery rhyme, but an approach that sees proteins 'twinkle' like stars in the night sky is providing new insight into an important class of enzymes involved in disease.
By tagging the enzymes with fluorescent dyes, researchers at Imperial have been able to reveal new characteristics, which they say could help to develop better drugs for parasitic infections, inflammatory disease and cancer.
The enzymes, called protein kinases, help to regulate cells and coordinate their responses to what's going on around them in the body. They are hugely important in cell signalling and raising the alarm when the cell's DNA is damaged, kicking off the response.
However, when the enzymes go wrong they can cause a whole host of problems downstream – such as unchecked growth, leading to tumours – which has made them an attractive target for drug makers developing new cancer treatments.
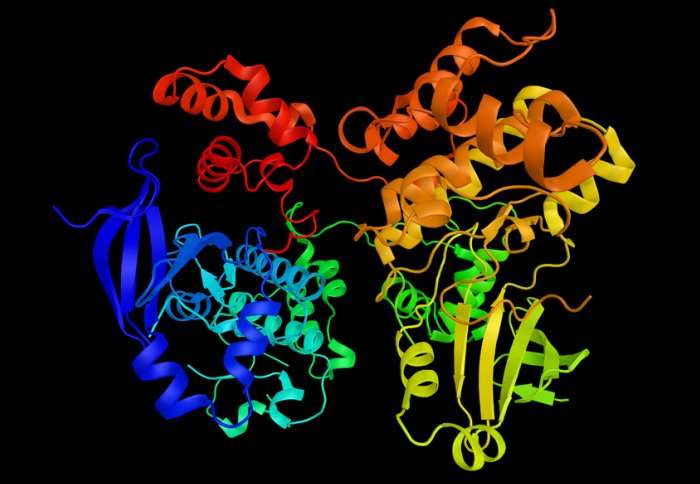
Previous research into their structure has revealed that the enzymes have 'active' and 'inactive' forms, depending on the shape and position of an 'activation loop'. Beyond this, however, it was not possible to measure how the enzymes switched between states, or if they needed a molecule to convert, and it could be hard to stabilise their state.
Now, a group led by Dr Charlotte Dodson, a research fellow in Imperial's National Heart and Lung Institute, has found a way to show the proportion of active and inactive forms and how the enzyme changes in response to molecules and inhibitors.
"We have known for about 15 years from crystal structures that these enzymes broadly fall into two classes structurally," explained Dr Dodson. "In the active form, the enzyme is 'ready to go' and to carry out its biological function, but in its inactive form it is unable to do its job."
Switching states
Working in collaboration with the Institute of Cancer Research, Dr Dodson's lab was able to tag fluorescent markers to two different regions of the enzyme.
When the enzymes were in the active state the dye molecules were far apart and fluoresced, appearing as a glowing dots under the microscope. When the enzymes switched to the inactive state, the two regions tagged with the dye came into close contact, quenching the dye and making the fluorescence twinkle out.
Depending on whether they added molecules that promoted active or inactive states, the team found they could alter the proportion of states in a sample, and they were able to measure it as a change in fluorescence.
"By measuring fluorescence, we can tell which form the kinases are in," said Dr Dodson. "When we do this continuously over time and look at individual molecules down a microscope, it looks like a night sky with lots of twinkling little dots.
"Because we can measure the fluorescence of individual molecules, we know they are changing in solution, and this is something no one has been able to do before."
According to the researchers, in practice, the approach could be used to help refine drugs that target the enzymes by showing how they affect the mix of active and inactive states.
For example, if an inhibitor pushed a sample of kinases to 70 per cent inactive 30 per cent active, drug makers could tweak the molecule and measure the impact on a population of kinases. If after tweaking the drug the mix changes to 90 per cent inactive, it would show the inhibitor is having a larger effect than previously.
Cancer is currently the largest target for kinase inhibitors, with a market estimated to be worth $30bn (USD) or more, but researchers are exploring the role of kinases in other areas, including diseases of the heart and lungs.
Dr Dodson added: "We've developed the test and carried out a proof of principle in one particular kinase to show that it works and to explore what's going on. The next step is to take things on and apply this to other kinases of interest.
"I think that as more and more people realise these enzymes are important in other diseases, the knowledge gained from oncology can be transferred across to new therapy areas."
She added: "The more information we can provide at the design stage of drug development, the better. By using this approach we could enable drug makers to design new treatments more efficiently, which could save time and money. It could lead to new inhibitors making it to the clinic faster."
More information: James A. H. Gilburt et al. Dynamic Equilibrium of the Aurora A Kinase Activation Loop Revealed by Single-Molecule Spectroscopy, Angewandte Chemie International Edition (2017). DOI: 10.1002/anie.201704654
Journal information: Angewandte Chemie International Edition
Provided by Imperial College London