August 8, 2017 report
Folktale diffusion traced using genomic data

(Phys.org)—A team of researchers with members from several European countries has conducted a study involving tracing the spread of common folktales throughout history in Eurasia. In their paper published in Proceedings of the National Academy of Sciences, the team describes using genome data to trace two common means of folktale distribution.
Most people today in the Western world are familiar with a handful of folktales, including "Hansel and Gretel," "Sleeping Beauty," "Cinderella" and "Rumpelstiltskin"—such tales typically have a moral or lesson. Those that struck a chord tended to be widely told and were passed down through the generations, first orally, then through books. The researchers with this new effort wanted to know whether such tales were distributed through the grapevine, so to speak, or whether they were carried by people moving from one place to another. To find out, they used some of the growing amount of publicly available genome data.
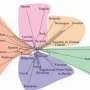
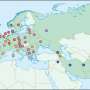
For their study, the researchers made a list of what they deemed the 596 most famous folktales in Europe and Asia—then, they compiled another list containing titles and information about published folktales. Next, they extracted information from global genome databases that provided data regarding the movement of people over different time periods. Connecting the two types of data allowed the researchers to create flow charts that described the movement of folktales over time.
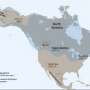
The researchers were able to see that both types of distribution were involved in the spread of folktales. Some of the tales moved through populations until they reached a border, either physical or social, such as a language barrier. Others were able to make giant leaps as people traveled great distances, taking the tales with them and relating them to those they encountered. The team notes that they were also able to isolate approximately 15 of the tales that had clearly spread due to migration. The researchers report that they were also able to narrow down the origination sites of some common fables to regions as broad as Northern Africa or Central Asia.
More information: Inferring patterns of folktale diffusion using genomic data, Eugenio Bortolini, PNAS, DOI: 10.1073/pnas.1614395114
Abstract
Observable patterns of cultural variation are consistently intertwined with demic movements, cultural diffusion, and adaptation to different ecological contexts [Cavalli-Sforza and Feldman (1981) Cultural Transmission and Evolution: A Quantitative Approach; Boyd and Richerson (1985) Culture and the Evolutionary Process]. The quantitative study of gene–culture coevolution has focused in particular on the mechanisms responsible for change in frequency and attributes of cultural traits, the spread of cultural information through demic and cultural diffusion, and detecting relationships between genetic and cultural lineages. Here, we make use of worldwide whole-genome sequences [Pagani et al. (2016) Nature 538:238–242] to assess the impact of processes involving population movement and replacement on cultural diversity, focusing on the variability observed in folktale traditions (n = 596) [Uther (2004) The Types of International Folktales: A Classification and Bibliography. Based on the System of Antti Aarne and Stith Thompson] in Eurasia. We find that a model of cultural diffusion predicted by isolation-by-distance alone is not sufficient to explain the observed patterns, especially at small spatial scales (up to ∼∼4,000 km). We also provide an empirical approach to infer presence and impact of ethnolinguistic barriers preventing the unbiased transmission of both genetic and cultural information. After correcting for the effect of ethnolinguistic boundaries, we show that, of the alternative models that we propose, the one entailing cultural diffusion biased by linguistic differences is the most plausible. Additionally, we identify 15 tales that are more likely to be predominantly transmitted through population movement and replacement and locate putative focal areas for a set of tales that are spread worldwide.
Journal information: Proceedings of the National Academy of Sciences
© 2017 Phys.org