Four-billion-year-old 'fossil' protein resurrected in bacteria protects them from viruses
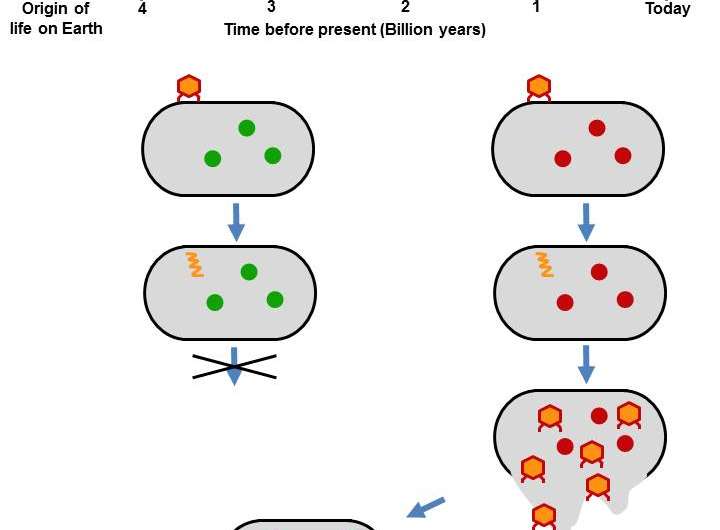
In a proof-of-concept experiment, a 4-billion-year-old protein engineered into modern E. coli protected the bacteria from being hijacked by a bacteria-infecting virus. It was as if the E. coli had suddenly gone analogue, but the phage only knew how to hack digital. The ancient protein, an ancestral form of thioredoxin, was similar enough to its present-day analogues that it could function in E. coli but different enough that the bacteriophage couldn't use the protein to its advantage. The work, which could be useful in plant bioengineering, appears May 9 in Cell Reports.
"This is an arms race. Thioredoxin has been changing in evolution to avoid being hijacked by the virus, and the virus has been evolving to hijack the protein," says senior author Jose Sanchez-Ruiz of the University of Granada in Spain. "So we go back, and we spoil all of the virus' strategy."
Sanchez-Ruiz's lab specializes in reconstructing ancient gene sequences that code for proteins. Since proteins do not preserve for billions of years, the researchers make their best estimation of the ancient protein based on genetic data across many different taxa. Thioredoxin, a versatile work-horse protein that moves electrons around so that chemical reactions in the cell can occur, is a favorite in the lab because it has been around almost since the origin of life and it is present in all modern organisms. We can't live without it, nor can E. coli.
Thioredoxin also happens to be one of the proteins that bacteriophage must recruit to survive and replicate. Without a hijack-able thioredoxin, the virus hits a dead end. In a series of experiments led by Asunción Delgado, then a post-doc at the University of Granada, the researchers tested seven reconstructions of primordial thioredoxins, ranging in age from 1.5 billion years old to 4 billion years, to see if they could function in modern E. coli.
The old-school thioredoxins passed the test with varying degrees of success. "That was a bit surprising," says Delgado. "The modern organism is a completely different cellular environment. Ancestral thioredoxins had different molecular partners, different everything. The farther back we get from present, the less they work in a modern organism. But even when we get back to close to the origin of life, they still show some functionality."
But the ancestral thioredoxins were just different enough that the modern phage couldn't recognize or bind to them.
However, resurrecting ancient proteins may be useful as more than a scientific curiosity. Virologists tend to focus on the human-infecting ones, but the viruses that kill the most people are not human pathogens but rather the viruses that kill off crops, sparking famines and mass starvation. Delgado, Sanchez-Ruiz, and their colleagues speculate that ancient proteins could be edited into plants to confer protection against crop-killing viruses. However, this idea has yet to be tested in plants.
"If this is applied to plants, it wouldn't be genes from ancient bacteria; it would be genes from the same plant. It would be the ancestral version of a gene from the same plant," says Sanchez-Ruiz. "This is genetic alteration, of course, but it is a mild genetic alteration. This is not like having a gene from one species being transferred to a different species. Also, this would not be like Jurassic Park. It would just be a comparatively small change in a gene that the plant already has."
Protein resurrection experiments could also shed light on how evolution works at the protein level. "What we can do is let the virus evolve to adapt to the ancestral protein, and then do the experiment in reverse," says Sanchez-Ruiz. "Once it's adapted to the ancestral protein, we can test how it reacts to the modern protein. We can see if it repeats the evolution. So it would be kind of a molecular version of this Stephen J. Gould 'replaying the tape of life' idea."
The researchers' next set of experiments will focus on the fundamentals of protein evolution, but they point out that understanding and resurrecting old proteins could be a key resource for biotech. Instead of introducing new elements, bioengineers may be able to re-use older ones from earlier in viruses and cells' co-evolutionary history. "Some people think that evolution is just a theory or is just some kind of philosophic explanation," says Sanchez-Ruiz. "Evolutionary studies have practical applications."
More information: Cell Reports, Delgado et al.: "Using Resurrected Ancestral Proviral Proteins to Engineer Virus Resistance" www.cell.com/cell-reports/full … 2211-1247(17)30531-4 , DOI: 10.1016/j.celrep.2017.04.037
Journal information: Cell Reports
Provided by Cell Press



















