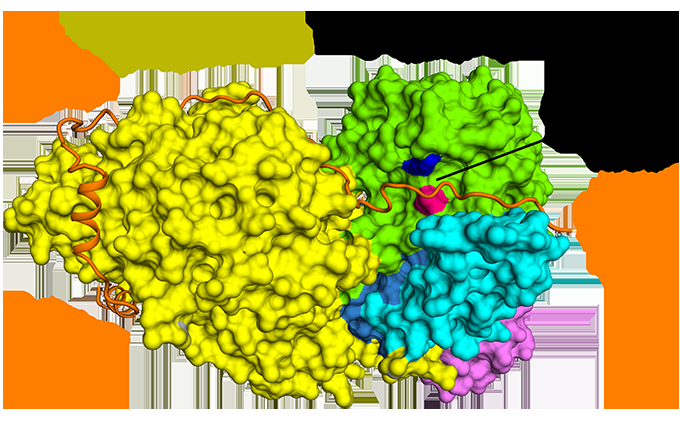
Figure 1: The antiparallel binding of securin compared to separase. Credit: Diamond Light Source
The structure of an important protein complex that regulates the metaphase-to-anaphase transition during cell cycle progression has been solved using cryo-electron microscopy (cryo-EM) at Diamond Light Source. The structural study, detailed in Nature Structural and Molecular Biology, has provided valuable insights into the inner workings of the separase–securin complex.
Separase is an enzyme that is responsible for cleaving the kleisin subunits (Scc1 and Rec8) of the cohesin ring that holds sister chromatids together during mitosis. Once the chromatids are liberated by separase, they segregate towards opposite poles of the cell, ready to form new nuclei in two identical daughter cells. Separase is kept in check by an inhibitory chaperone known as securin, which is intriguingly also believed to have activating properties.
Although discovered almost 20 years ago, it is only recently that the structure of separase bound to securin has been elucidated. Cryo-EM was undertaken at the Electron Bio-Imaging Centre (eBIC) at Diamond to determine the structure of the full complex and to explore the molecular interaction between separase and securin. It was discovered that securin forms an extended conformation to interact along the entire length of separase, and inhibits the enzyme through a pseudosubstrate mechanism at the active site. A full understanding of this interaction and nature of cell cycle control could open up new avenues for targeted drug design.
A cell cycle regulatory complex
When chromosomes are replicated they each contain two sister chromatids, and during cell division these chromatids must separate in a coordinated and accurate manner to produce two identical daughter nuclei. At the beginning of mitosis, the sister chromatids are held together by a protein complex called cohesin; however, during the metaphase-to-anaphase transition cohesin is cleaved by a protease enzyme known as separase to separate the chromatids and allow them to move to opposite sides of the cell just before it divides.
The transition of metaphase to anaphase begins when a large multi-subunit E3 ubiquitin ligase, called the anaphase-promoting complex (or APC/C) marks an inhibitory chaperone, called securin, with ubiquitin for destruction. Once securin is degraded, separase is free to cleave cohesin and separate the chromatids.
The separase–securin complex is critical for cell division, yet the structure of this duo had not been solved in its entirety. Dr Andreas Boland, Postdoctoral Research Fellow in Dr David Barford's group at the MRC Laboratory of Molecular Biology and co-lead investigator of the study explained why this complex is so fascinating: "The mis-segregration of genetic material can lead to aneuploidy and tumorigenesis, therefore this is one of the most important steps during cell cycle progression. Paradoxically, securin not only prevents premature cleavage of cohesin, but it can also promote separase activity. To understand the dual mechanisms of activation and repression in detail we used single-particle cryo-EM."
Cryo-EM structure at 3.8-Å resolution
Separase–securin complexes from both C. elegans and humans were expressed, purified, and applied to graphene-oxide-covered gold carbon grids before being flash frozen in liquid ethane. Cryo-EM micrographs were collected using the Titan Krios microscope at eBIC with a calibrated magnification of 47,619, which corresponded to a pixel size of 1.05 Å.
Dr Boland noted the value of cryo-EM for this application: "The protein complexes are relatively small in size, with less than 250 kDa for the human and only around 150 kDa for the C. elegans complex, but they contain multiple domains and are flexible, so they were challenging materials to crystallise. Cryo-EM is a feasible option even for small protein complexes like this."
For the first time ever, cryo-EM was used to determine the near-atomic resolution structure of the C. elegans separase–securin complex. Furthermore, the structure of the human equivalent of the complex was solved to a medium resolution, revealing the evolutionary conservation of separase's triangular shape.
Cryo-EM allowed the team to visualise how securin engages with separase. It adopts an extended anti-parallel conformation to bind onto hydrophobic patches along the entire length of separase. Curiously, these hydrophobic interactions were also shown to stabilise separase, potentially explaining the activating effects of securin on separase. Moreover, securin interacts with the catalytic site of separase via a pseudosubstrate mechanism. An arginine residue present at the P1 cleavage position in the cohesin substrates Scc1 and Rec8 is replaced in securin by a hydrophobic residue. This substitution is responsible for distorting the geometry of the separase catalytic site at the substrate cleavage position, consequently blocking its protease activity.
Separase is crucial for cell cycle progression, so it could be an interesting pharmaceutical target. Mutations, overexpression, and mislocalisation of separase have been shown to be associated with tumour developments such as breast cancer.
Dr Boland plans to continue studying the complex: "In future, we want to improve the resolution of the human separase–securin complex to achieve an atomic or near-atomic resolution. We would also like to understand how the separase-securin complex is bound and ubiquitinated by the APC/C to mark it for destruction, so we will continue using cryo-EM for this purpose."
More information: Andreas Boland et al. Cryo-EM structure of a metazoan separase–securin complex at near-atomic resolution, Nature Structural & Molecular Biology (2017). DOI: 10.1038/nsmb.3386
Journal information: Nature Structural and Molecular Biology , Nature Structural & Molecular Biology
Provided by Diamond Light Source