Accurate DNA misspelling correction method
Researchers at the Institute of Basic Science (IBS) proved the accuracy of a recently developed gene editing method. This works as "DNA scissors" designed to identify and substitute just one nucleotide among the 3 billion. "It is the first time that the accuracy of this base editor has been verified at the whole genome level," explains KIM Jin-Soo, leading author of the study. Published in Nature Biotechnology, this validation will help to expand the use of this method in agriculture, livestock, and gene therapy.
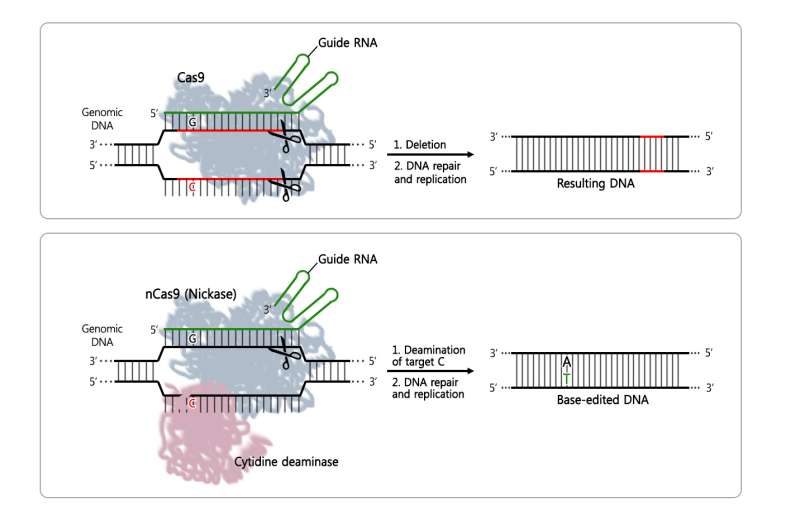
Rapid advances in gene editing tools have created excitement in the biology community. The primary third-generation DNA editing technology is CRISPR—a tool that is quicker and cheaper than its predecessors. By cutting out a small DNA sequence, CRISPR-Cas9 and CRISPR-Cpf1 are used to silence or reduce the expression of faulty genes. However, last year, biologists discovered a new base editor method that does not cause random DNA deletions and insertions, but instead replaces only one DNA base. These types of gene corrections are critical, as several diseases are caused by the misspelling of one of the four basic components of DNA; adenine (A), cytosine (C), guanine (G), and thymine (T). Single-nucleotide errors in DNA are referred to as point mutations. Examples of conditions caused by point mutations include cystic fibrosis, sickle cell anemia and color blindness.
Unlike the existing technologies, the base editor method consists of a variation of CRISPR-Cas9 (nCas9, nickase) fused with another enzyme called cytosine deaminase, which replaces the DNA component C with T. The scissors are directed to the correct position on the DNA by a guide RNA. However, until recently, it was not known whether the base editor was working only in the area of the faulty gene or if it was unnecessarily substituting Cs in off-target areas.
Just one month after reporting the first successful base editing in animals in Nature Biotechnology to modify a single nucleotide in dystrophin and tyrosinase genes, the same team demonstrated the accuracy of this method at the genome scale.
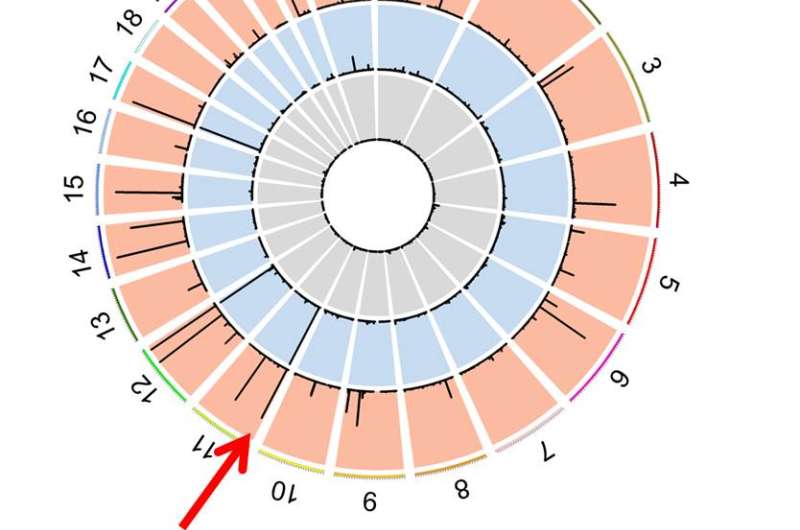
In order to identify the soundness of the method, IBS researchers modified the error-checking technique known as Digenome-seq, in order to adapt it to the base editor method. Digenome-seq was used and validated last year, when the team analyzed the accuracy of CRISPR-Cpf1 and Cas9. IBS researchers also improved the computer program Digenome 2.0 to identify off-targets more comprehensively and compared different guide RNAs to find the one that reduces malfunctions and increases specificity.
Using this technique, the team found the base editing technique to be even more accurate than the current third-generation CRISPR-Cas9. The base editing technique induced C-to-T conversions in multiple sites in the human genome, while CRISPR-Cas9 caused cleavages in multiple sites, meaning that the new base editor technique makes fewer off-target changes. "Therefore, it is expected that these base editors will be used as widely as the popular CRISPR technology," says KIM.
More information: Genome-wide target specificities of CRISPR RNA-guided programmable deaminases, Nature Biotechnology (2017). nature.com/articles/doi:10.1038/nbt.3852
Journal information: Nature Biotechnology
Provided by Institute for Basic Science