Team finds new way that bacteria lay siege to neighboring cells by hijacking two factors involved in protein synthesis
So they can't use smartphones or WiFi, but bacteria have evolved some seriously complex strategies to communicate with one another. And the resulting interactions are a delicate balance of cooperation and, in some cases, competition.
These intraspecies exchanges take place within contact-dependent growth inhibition (CDI) systems, which regulate cellular activities via cell-to-cell contact and are found in a wide variety of gram-negative bacteria, including important human pathogens such as Escherichia coli.
New research by UC Santa Barbara biologists examines how a particular pathogenic strain of E. coli—EC869, which causes diarrhea or hemorrhagic colitis in humans—destroys its neighbors by transferring toxins that inhibit their cell growth. Previous work by other UCSB researchers had shown that a different variation of E. coli required a "permissive factor" for toxin activation. The authors of the new paper, which appears in the Proceedings of the National Academy of Sciences, wondered whether EC869 also needed to bind protein in the target cell to activate its toxin.
The answer is yes, with a twist.
Many toxins inhibit translation, the essential process by which the sequence of messenger RNA (mRNA) is converted into protein. Some toxins accomplish this by breaking transfer RNAs (tRNAs), molecules that help decode mRNA sequences, lead author Allison Jones noted.
"We discovered that a class of these tRNA-cleaving poisons intoxicates cells differently from previous toxins in that they hijack two essential factors involved in protein synthesis and use them to find their cellular victims," said Jones, a graduate student in UCSB's Department of Molecular, Cellular and Developmental Biology (MCDB).
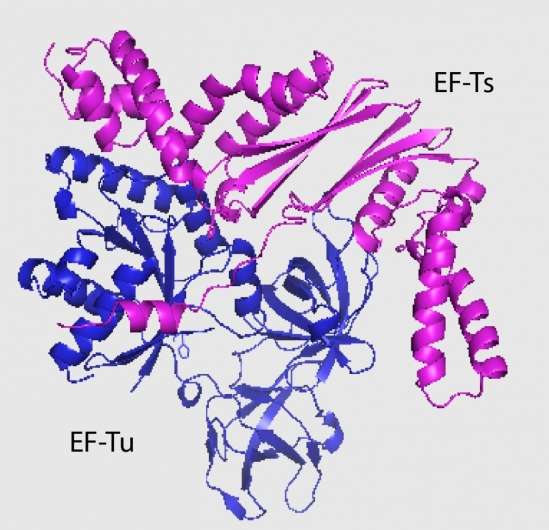
Jones and her colleagues discovered that the EC869 toxin binds to the most abundant protein in bacteria, elongation factor Tu (EF-Tu). EF-Tu's role in protein synthesis is to bind to tRNA molecules. The binding action not only activates toxins but also helps zero in on the true target: tRNA molecules. And not just any tRNA: EC869 cleaves only two specific tRNA molecules out of 46 types present in the cell.
"It appears that these toxins are riding piggyback on EF-Tu to find tRNAs and seem important for their own stabilization in the cell as well," said co-author Fernando Garza-Sánchez, a MCDB staff research in the Hayes Lab. "However, in addition to EF-Tu, these toxins also require the presence of another elongation factor, EF-Ts."
"We were unsure of the role of EF-Ts, as the toxin does not appear to bind stably to it," Jones explained.
A possible hint comes from the fact that this class of toxins only cuts specific tRNA molecules. The researchers hypothesize that the toxins sit bound to EF-Tu as EF-Ts loads and unloads different tRNA molecules until the specific target arrives. Once the right tRNA is loaded, it is cleaved, destabilized and released.
"Our research lends support to an unestablished role for EF-Ts—that of having an active role in delivering tRNAs to EF-Tu during protein synthesis," Jones said. "It will be interesting to see if the interaction of this toxin with members of the protein synthesis apparatus plays a role in intercellular communication."
More information: Allison M. Jones et al. Activation of contact-dependent antibacterial tRNase toxins by translation elongation factors, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1619273114
Journal information: Proceedings of the National Academy of Sciences
Provided by University of California - Santa Barbara