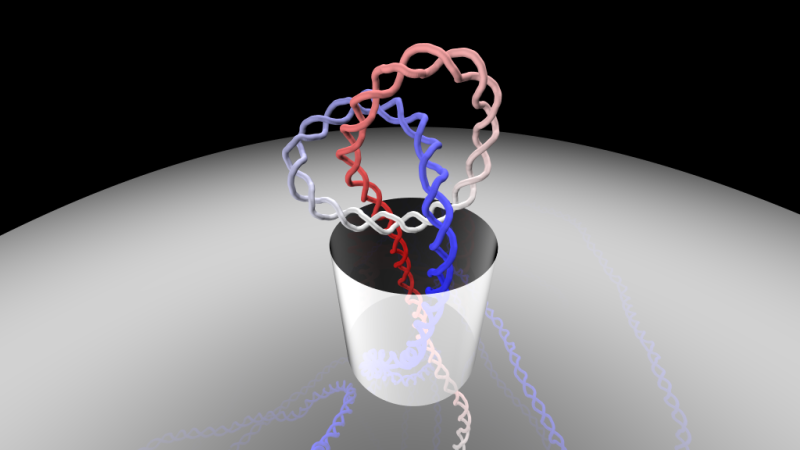
How can long DNA filaments, which have convoluted and highly knotted structure, manage to pass through the tiny pores of various biological systems? This is the fascinating question addressed by Antonio Suma and Cristian Micheletti, researchers at the International School for Advanced Studies (SISSA) in Trieste who used computer simulations to investigate the options available to the genetic material in such situations. The study has just been published in PNAS, the journal of the National Academy of Sciences of the Unites States. Credit: Antonio Suma, SISSA
Anyone who has been on a sail boat knows that tying a knot is the best way to secure a rope to a hook and prevent slippage. Similarly, knots in sewing threads prevent them slipping through two pieces of fabric. How, then, can long DNA filaments, which have convoluted and highly knotted structure, manage to pass through the tiny pores of various biological systems? This is the fascinating question addressed by Antonio Suma and Cristian Micheletti, researchers at the International School for Advanced Studies (SISSA) in Trieste, who used computer simulations to investigate the dynamics of the molecule in such situations. The study has just been published in PNAS, the journal of the National Academy of Sciences of the Unites States.
"Our computational study sheds light on the latest experimental breakthroughs on knotted DNA manipulation, and adds interesting and unexpected elements," explains Micheletti. "We first observed how knotted DNA filaments pass through minuscule pores with a diameter of about 10 nanometers (10 billionths of a meter). The behaviour observed in our simulations was in good agreement with the experimental measurements obtained by an international research team led by Cees Dekker, which were published only a few months ago in Nature Biotechnology. These advanced and sophisticated experiments marked a turning point for understanding DNA knotting. However, current experiments cannot detect how DNA knots actually pass through the narrow pore.
"In fact, the phenomenon occurs over a tiny spatial scale that is inaccessible to microscopes. This is why our group resorted to what the great German biophysicist Klaus Schulten called 'the computational microscope,' that is, computer simulations."
Suma and Micheletti explain: "The simulations revealed that the passage of the knot can occur in two distinct ways: One where the knot is tight, and the other where the knot is more delocalised. In both cases, the knot not only passes through the pore, but it does so in a very brief time."
Moreover, the knot usually passes in the final stages of the translocation, when most of the DNA strand has already passed. "But there is something more that is counterintuitive," state the authors. "The size of the knot, whether small or large, does not seem to affect the pore obstruction time by much. The latter depends instead on the translocation speed, which, in turn, depends on the initial position of the knot along the filament." These results, say the researchers, ought to help the design of future experiments probing the spontaneous knotting of DNA, a still largely unexplored venue, especially regarding the size of DNA knots.
A short video of the computer simulation. Credit: Antonio Suma, SISSA
Advancing our current understanding of knots in biological molecules is important to clarify their implications in biological contexts as well as in applicative ones, such as DNA sequencing using nanopores. Suma and Micheletti hope that the promising directions suggested by their study can lead to a more detailed and accurate profiling of entanglement in DNA, RNA and proteins.
More information: Antonio Suma et al. Pore translocation of knotted DNA rings, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1701321114
Journal information: Proceedings of the National Academy of Sciences , Nature Biotechnology
Provided by Scuola Internazionale Superiore di Studi Avanzati