Novel mechanism that detains mobile genes in plant genome

A team of Hokkaido University researchers has discovered a hitherto-unknown mechanism that detains transposable elements or "mobile genes" - which can move and insert into new positions in plant genomes.
Transposable elements (TEs), also known as mobile genes, are considered parasites of host genomes because they act as powerful mutagens. If not kept in check, they can cause gene disruption, genome rearrangement and genomic takeover. Thus, an essential function of organisms is controlling the movements of this troublemaker. Until now, all identified TE regulations were epigenetic-dependent, meaning that the production of TE proteins are suppressed.
A TE called Tam3 in snapdragons (Antirrhinum majus) can be regulated into active and inactive states through temperature fluctuations. It is thus possible for researchers to identify a mechanism whereby TE falls into an inactive state. The Hokkaido University research team then focused on the Tam 3 transposase protein, which is produced by the TE to enable it to move, and by employing various means investigated its positions in the cell.
According to their research, snapdragons detained Tam3 transposase within the plasma membrane when Tam3 was inactivated. When Tam3 was activated, Tam3 moved to the cell nucleus, where it is normally found.
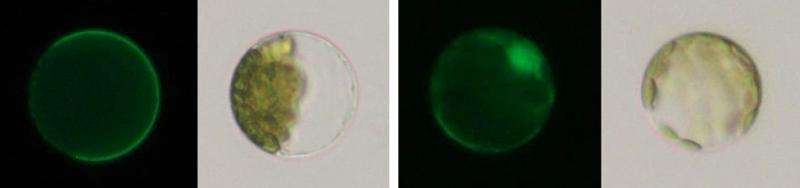
The team also found that a structure called "Znf-BED" within Tam3 transposase plays a pivotal role in detaining Tam3 at the plasma membrane. When part of Znf-BED was changed, the transposase did not move to the plasma membrane and instead entered the cell nucleus. The team thus suggests that unknown protein produced by snapdragons binds to Tam3 transposase through Znf-BED, and detains them at the plasma membrane.
"It is the first time that such a TE detainment has been discovered," says Kaien Fujino in the research team. "The newly-found mechanism, which detains TEs after proteins are produced, is different from epigenetic regulation, where gene expression is controlled before protein is generated. Our findings should facilitate research on similar mechanisms of mobile elements in other organisms."
More information: Hua Zhou et al. Detainment of Tam3 Transposase at Plasma Membrane by Its BED-Zinc Finger Domain, Plant Physiology (2017). DOI: 10.1104/pp.16.00996
Provided by Hokkaido University