New study shows that proteins are 'virtually' knotted
Many of the processes essential to life involve proteins - long molecules which 'fold' into three-dimensional shapes allowing them to perform their biological role.
Consisting of strings of amino acids, a folded protein molecule resembles a coiled, tangled piece of wire, which, as everyday experience suggests, may be knotted.
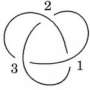
The mathematical study of knots is called knot theory, a branch of abstract mathematics which is related to other areas of maths such as algebra. The knotted curves studied in knot theory have closed ends, like a knot in a circle, but protein molecules do not.
A new study by physicists at the University of Bristol has shown that knots in proteins can be understood using 'virtual knots' - a branch of knot theory previously considered as abstract and without application. Earlier research into knotted proteins involved adding lines to close a protein curve into a loop. As there is no obvious single way to do this, researchers took averages over many different closure lines.
Professor Mark Dennis, from the School of Physics, said: "Our procedure, however, takes views of the protein curve from different directions, that is, projections, which can be mathematically analysed as virtual knots without adding extra lines. This captures the essential ambiguity of where the ends of the protein curve are."
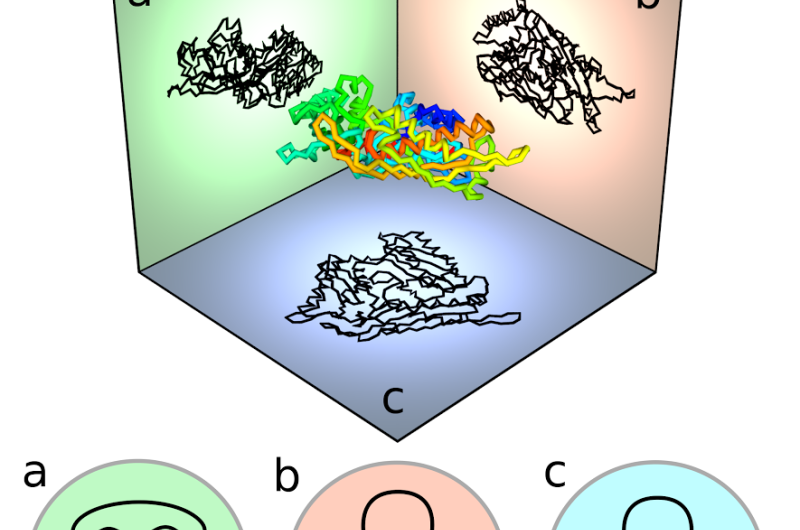
Viewing the protein curve in different directions results in different projections, or 'shadows', of the curve. The virtual knotting of each shadow can be identified mathematically from the sequence of over and under crossings of the projection.
The various types of virtual or regular knot that occur in each direction, not obvious without smoothing the projection, can be drawn on a spherical map; the 'globe' of viewing directions is broken up into 'seas and islands' of different knot types. The three-dimensional structure of the protein, essential to its function, can be better understood from the different kinds of map that appear in this way.
When the protein knots are closed by extra lines, the seas and islands are restricted to only a small number of 'classical' knot types—those of knotted circles. Since there are many more virtual knot types than classical types (as they don't have to close), viewing the knot 'virtually' offers a more subtle understanding about the protein molecule's shape.
This work is part of the Scientific Properties of Complex Knots (SPOCK) project, a collaboration between the University of Bristol and Durham University.
The aim of the project is to create new computational tools and mathematical techniques for the analysis, synthesis and exploitation of knotted structures in a wide range of complex physical phenomena.
More information: 'Proteins analysed as virtual knots' K. Alexander, A. J. Taylor and M. R. Dennis, Scientific Reports, 2017.
Provided by University of Bristol