New kit helps researchers make sense of mass cytometry datasets to uncover cell subsets
A new software package offers easier analysis and interpretation of experiments that use mass cytometry, a sophisticated method for determining the properties of cells. The tool—called cytofkit—enables scientists to identify different subpopulations of cells within a sample of immune cells, cancer cells or other tissue types.
Flow cytometry remains the go-to method for biological investigations that require single-cell resolution. But, because the technology relies on fluorescent tags to detect different markers within the cell, only a limited number of labels can be applied before the light signals start to bleed into one another.
Mass cytometry helps solve this problem. By using metal labeling, the technique allows scientists to measure many more characteristics simultaneously within individual cells. But sorting through all the data it produces can be challenging, and most researchers agree that better analytic tools are needed.
Jinmiao Chen and her colleagues at the A*STAR Singapore Immunology Network made cytofkit in response to this need. The package combines state-of-the-art bioinformatics methods and in-house novel algorithms to help anyone make sense of mass cytometry data. "It provides a very user-friendly graphical interface and interactive visualization of analysis results," says Chen. "Anybody, including bench scientists and non-bioinformaticians, can use it without any training."
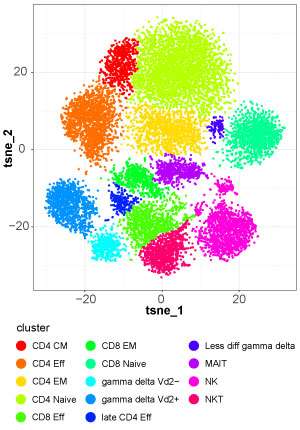
The software involves four main steps: first, cytofkit performs data pre-processing according to the users' specifications; second, the software automatically identified different matching subsets of cells; third, it allows visualization of the data with color-labeled cell types; and lastly, it infers the relatedness between cell groups.
Chen's team tested the tool's performance on mass spectrometry results collected from a sample of white blood cells. As they reported in PLOS Computational Biology, the software correctly identified known subpopulations of cells and further segregated these subsets to reveal additional cell types. In collaboration with A*STAR immunologist Evan Newell, the researchers also showed that cytofkit revealed many types of follicular helper T cells from blood and tonsils. Plus, says Chen, "we have tested the utility of cytofkit on a large number of other datasets not mentioned in the paper."
Cytofkit is also gaining popularity with scientists around the world. "It now has more than 4,000 users," says Chen. Her lab continues to improve and upgrade the tool in response to user feedback.
The software—which works both on flow and mass spectrometry datasets alike—is freely available through Bioconductor, an open-source software framework for biologists. It can be found here.
More information: Hao Chen et al. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline, PLOS Computational Biology (2016). DOI: 10.1371/journal.pcbi.1005112
Journal information: PLoS Computational Biology