Towards therapeutic applications of mRNA—new insights into translation and decapping
Gene therapy gives hope to millions of patients. Researchers from the University of Warsaw have been working on mRNA containing a modified fragment that initiates protein biosynthesis. Recently published results reveal that new compounds designed and synthesized at the University of Warsaw are more stable and effective than their natural equivalents, and their synthesis is simpler. The compounds lend a better understanding of the mechanisms of protein biosynthesis in cells, which in turn could lead to better therapeutics.
Protein production is frequently disrupted in the cells of a disease-affected organism. This manifests as imbalance in the synthesis of certain proteins or production of damaged proteins, which in extreme cases leads to cancer. Gene therapy is one approach to this problem. It involves supplying the organism with genetic material encoding proteins whose properties support healthy cell activity. In the early days of experimental gene therapy, researchers used DNA as the genetic material. However, genes delivered in the form of DNA integrate with the patient's genome, which may cause serious and unexpected symptoms. Medical researchers have high hopes about the therapeutic potential of mRNA; the molecules are smaller and simpler, which makes them easier to prepare under laboratory conditions, and perhaps most importantly, unlike DNA, they don't make permanent changes to the organism's genetic makeup.
mRNA molecules are natural polymers formed in cells. They contain precise copes of genes (DNA fragments), so they carry the genetic code and act as templates in the production of new proteins. The mRNA molecules are broken down by enzymes after a few minutes or hours. This short lifespan of natural and synthetic mRNA is one of the problems limiting practical applications. The use of mRNA in gene therapy would be more feasible if the molecule used in drugs survived for longer than its natural equivalent, and if the therapeutic efficacy was as high as possible.
In 2011, the consortium between the University of Warsaw and the Louisiana State University patented and commercialized the team's invention, improving the stability and efficiency of mRNA. The solution is currently undergoing clinical tests conducted by one of the pharma partners. The key innovation is the five-prime cap (5' cap), an artificial mRNA segment replacing its natural 7-methylguanosine structure.
The ongoing Warsaw research aims to discover improved cap analogues, design a technology for large-scale production of therapeutic mRNA, and improve understanding of the course of natural protein synthesis. "The 7-methylguanosine cap is at the 5' end of the mRNA molecule," explains Prof. Darzynkiewicz. "In cytoplasm, the cap structure is recognized by the eIF4E factor, which initiates the process of protein biosynthesis, known as translation. This stage decides on the speed of the entire complex sequence of events, which culminates with the synthesis of protein in the cell. The cap protects the mRNA from degradation by cleaving enzymes—nucleases. Unfortunately, cells remove the cap using decapping enzymes such as Dcp1/Dcp2. A few years ago, we discovered that using modified cap analogues can prevent the degradation of the 5' mRNA end, and improve the rate of translation."
The team reports that the search for natural cap analogues is promising. "Our recent paper, published in Nucleic Acids Research, presents a new class of modified caps which are an improved version of those currently undergoing clinical trials," says Prof. Jemielity. "The modification involves swapping an oxygen atom for a sulfur atom in several positions in a specific place of the cap molecule, known as the tri- or tetra-phosphate bridge. mRNA with this chemically modified cap is bound effectively by the eIF4E factor during the stage limiting the speed of protein biosynthesis. It's also highly resistant to the cleaving of the cap structure by the Dcp1/Dcp2 enzyme. Under cellular conditions, this mRNA is more stable and produces higher amounts of therapeutic protein, which we have demonstrated in a model used in studies of cancer vaccines. We hope that our modified mRNA will enable us to use lower doses of therapeutics—and lower doses mean a lower risk of side effects."
The drug's availability is also very important for the patient. Traditional enzyme methods of modifying caps (and in turn therapeutic mRNA) are time consuming and ineffective. "Back in 2010, it took us six months to prepare the first four grams of cap needed to start clinical trials, and the amount was barely sufficient to treat 12 or 13 patients," recalls Dr. Kowalska. Meanwhile, the potential demand can be estimated at kilograms every year, leading researchers to seek faster and cheaper production methods.
"We turned our attention to click chemistry," says Sylwia Walczak, Ph.D. student at the University of Warsaw. "We have been developing a method of effective synthesis of cap analogues from prefabricated units—chemical 'building blocks'. The structure of each block has at least one fragment which joins its counterpart in another molecule, interlocking like bricks."
By applying the method in straightforward production of modified caps, scientists from Poland have developed 36 new analogues. "Two of the compounds have properties we were hoping for. When they are introduced to mRNA, they work as well as the natural cap," says Anna Nowicka, who is working on her Ph.D. at the University of Warsaw. "We are certain that this discovery will pave the way to developing new chemical methods of adding the cap to mRNA, which will compete with expensive and time-consuming enzymatic methods." The work was published in late summer in the journal Chemical Science.
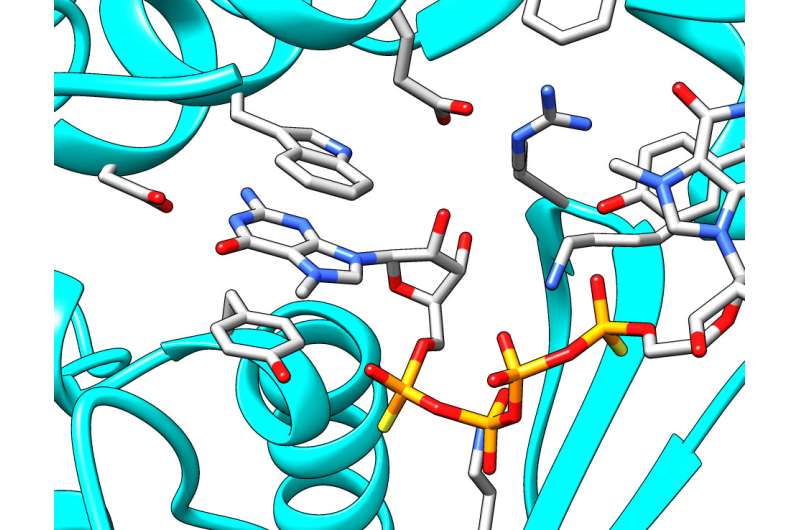
The search for new, improved cap analogues slowly shifts from the trial and error approach toward rational design. This is possible due to advances in the understanding of mRNA-related processes, their control and dynamics. Recently, the team from Warsaw contributed to new insights into mRNA decapping. "For the first time we have been able to design compounds which, by mimicking the 5' mRNA cap, are able to inhibit the Dcp1/Dcp2 enzyme, which cleaves the cap from mRNA exposing it to degradation," says Dr. Marcin Ziemniak, who completed his Ph.D. at the Faculty of Physics at the University of Warsaw earlier this year. "Working with colleagues at the University of California in San Francisco, John D. Gross and Jeffrey Mugridge, we have used X-ray crystallography to get new insight into structure and function of Dcp1/Dcp2. We have used our compound to capture the key stage of enzyme activity, which is binding the cap. To put it more simply, we used our compound as a bait, which imitates the mRNA cap. The enzymatic complex 'swallows' the bait, 'freezes,' and can be 'photographed'. Our results indicate that as the bait is taken—the inhibitor is bound—the enzyme complex undergoes global structural changes. The chemical composition of molecules remains unchanged, of course, but their fragments rotate relative to one another to reach a situation when the enzyme is ready to act."
The results have been published in two prestigious journals: RNA (January) and Nature Structural and Molecular Biology (October). "We believe that the results will allow us to design even better inhibitors of mRNA decapping," says Prof. Jemielity. "They will be useful in further research into mRNA degradation processes, and hopefully they will also find therapeutic applications such as increasing the potency mRNA-based gene therapies."
Scientists stress that the problems they are working on require an interdisciplinary approach. "The work we are conducting at the Faculty of Physics is unique," says Dr. Kowalska. "We have access to state-of-the-art research labs, although it's true to say that other teams have similar equipment. Our advantage lies in our team, which consists from experts in biophysics, chemistry and molecular and cellular biology. Conducting research on the boundaries of three different disciplines and the ability to look at the same research problem from different perspectives is incredibly inspirational, and gives us opportunity to come up with completely fresh ideas and solutions which would be far more difficult to reach using just a single approach. I believe this is a unique approach not only in Poland but on a global scale," Kowalska sums up the situation.
More information: Malwina Strenkowska et al, Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential, Nucleic Acids Research (2016). DOI: 10.1093/nar/gkw896
Sylwia Walczak et al. A novel route for preparing 5′ cap mimics and capped RNAs: phosphate-modified cap analogues obtained via click chemistry, Chem. Sci. (2017). DOI: 10.1039/C6SC02437H
Jeffrey S Mugridge et al. Structural basis of mRNA-cap recognition by Dcp1–Dcp2, Nature Structural & Molecular Biology (2016). DOI: 10.1038/nsmb.3301
Journal information: Chemical Science , Nucleic Acids Research , Nature Structural and Molecular Biology , Nature Structural & Molecular Biology
Provided by University of Warsaw