Predictive kinetic model paves the way for designing microbial factories
The microbe E. coli gets a bad rap in the food industry, but in the chemical engineering field, it is one of the most important microorganisms for producing amino acids, bioethanol, vitamins and more. Researchers in Penn State's Chemical and Biological Systems Optimization Lab have developed a near genome-scale kinetic model of Escherichia coli's metabolic processes, which will allow scientists to quickly and efficiently use computer modeling to predict the effects that multiple gene interventions have on the microbial factories.
"This could eventually be transformative in our ability to reliably design novel overproducing microbial strains," said Costas D. Maranas, Donald B. Broughton Professor of Chemical Engineering.
Maranas explained that recently developed techniques for changing the native metabolism of microbes allow for the manipulation of multiple genes at the same time, quickly and cost-effectively. When using microbes as factories for the production of chemicals or biofuels, the key is to find the best candidates out of hundreds of genes in order to harness the subsequent increases in productivity. The new kinetic models provide these results of metabolic changes to microbes like E. coli. "This opens the door to testing in the computer many combinations and find the best one before having to expend time and resources in the lab," Maranas said.
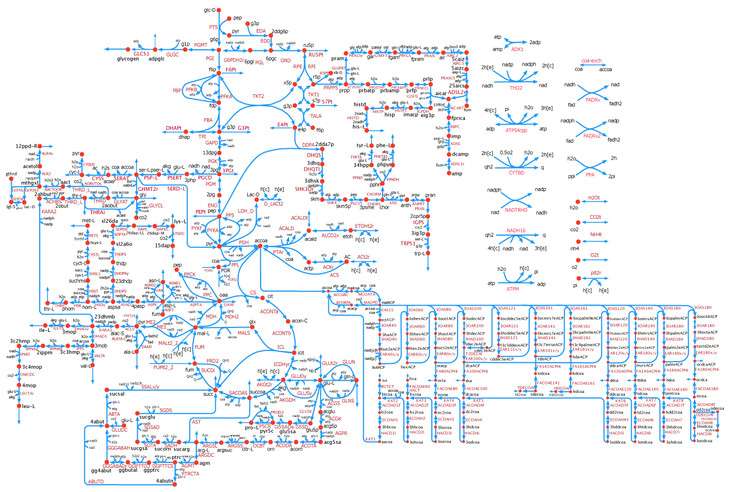
The model, called k-ecoli457, contains 457 model reactions, 337 metabolites, and 295 substrate-level regulatory interactions. Model k-ecoli457 was parameterized using metabolic flux data for both wild-type E. coli and 25 mutant strains. Comparisons of the k-ecoli457 model's predictions against multiple experimentally measured datasets showed substantial accuracy improvements over previous models.
These advances in designing novel strains aren't limited to E. coli; similar kinetic models could be developed for other microorganisms such as cyanobacterial or clostridia.
More information: The model is available for download at www.maranasgroup.com
Ali Khodayari et al. A genome-scale Escherichia coli kinetic metabolic model k-ecoli457 satisfying flux data for multiple mutant strains, Nature Communications (2016). DOI: 10.1038/ncomms13806
Journal information: Nature Communications
Provided by Pennsylvania State University