New tool to track circulating cancer cells
Cancerous tumours are known to release cells into the bloodstream, and it is these circulating tumour cells or CTCs that are the sources of metastatic tumours – tumours that spread and form in distant locations in the body. In fact, most patients who succumb to cancer do not die because of the initial tumours that form, but rather because of the deadly secondary metastatic tumours that appear at distant sites. As a result, understanding the biology and clinical relevance of these traveling cells is critical in our fight against cancer.
Monitoring circulating tumour cells, however, is a tremendous challenge as they are outnumbered in blood by healthy cells at a level of over 1 billion-to-1. Moreover, they can display varied and dynamic properties, and the collection of CTCs found in the bloodstream of a cancer patient may have differing metastatic potential. Consequently, efforts to integrate the analysis of these cells into mainstream clinical medicine have been limited because it has been difficult to pinpoint what types of cells and what phenotypic properties should be targeted. But the potential of CTCs to allow the collection of a non-invasive "liquid biopsy" to monitor cancer progression is a tantalizing possibility that has continued to attract significant attention to this problem.
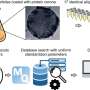
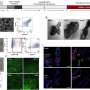
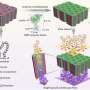
A breakthrough by Professor Shana Kelley's research group at the University of Toronto published in Nature Nanotechnology provides a new tool to characterize CTCs that may help cancer biologists and clinicians understand how to use these cells to provide better treatment. Using magnetic nanoparticles, CTCs in blood samples were targeted based on proteins displayed on the cell surface, and separated based on the levels of the protein present. Using a high–resolution microfluidic device, cells are then separated into 100 different capture zones to generate a profile that provides phenotypic information related to metastatic potential. Using this approach and monitoring cells generated in animal models of cancer and in samples collected from prostate cancer patients, the properties of CTCs were shown to evolve and become more aggressive as tumours became more advanced.
"Through this approach, we aimed to provide a new way to profile CTCs beyond simply counting their numbers in clinical samples," explained Dr. Mahla Poudineh, lead student author on the paper. "Instead, we wanted to provide phentotypic information that might allow these cells to be classified as benign or more dangerous, which would then inform treatment options."
"We were very fortunate to collaborate with a number of oncologists at the Sunnybrook Research Centre and Princess Margaret Hospital as we developed this technology so that we could test our approach with real patient specimens and better understand how to adapt it for use in the clinic," noted Dr. Kelley.
The Kelley group (www.kelleylaboratory.com/), along with collaborators in the Sargent group (www.light.utoronto.ca/) at the University of Toronto, hope to turn the approach they reported into a device that can be used by cancer researchers and eventually clinicians to allow CTC analysis to be monitored routinely and used to limit the progression of cancer.
More information: Mahla Poudineh et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking, Nature Nanotechnology (2016). DOI: 10.1038/nnano.2016.239
Journal information: Nature Nanotechnology
Provided by University of Toronto