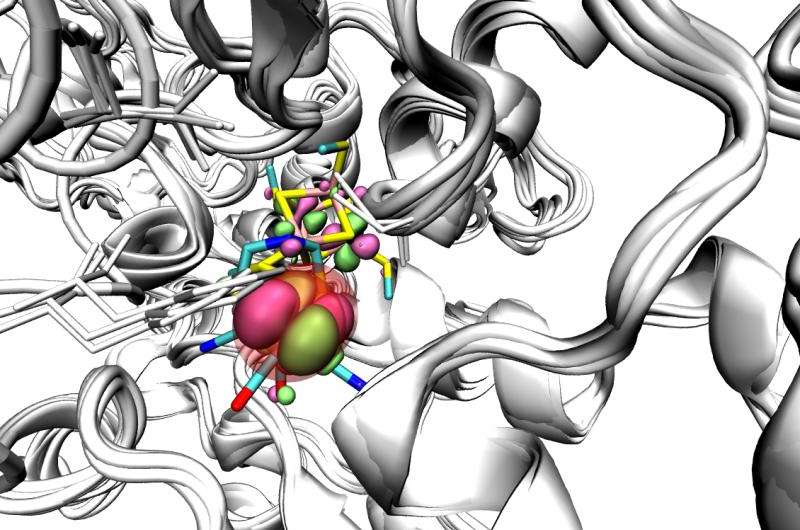
Visualization of the active site (coloured sticks) that is degraded by oxygen molecules within the core of the Fe-Fe hydrogenase with SOMO orbitals shown in green/pink. Credit: Elhuyar Fundazioa
Oxygen inhibits hydrogenases, a group of enzymes that are able to produce and split hydrogen. This degradation is fatal for possible biotechnological applications of these enzymes for the production of clean energy. Understanding the mechanisms of this process is thus essential. An international team lead by researchers from UCL (UK) and CNRS (France), including an Ikerbasque Research Fellow from CIC nanoGUNE, have combined theory and experiment to characterize each chemical reaction step that results in the reduction of oxygen by the enzyme. These results are being published this week in the journal Nature Chemistry.
Fossil fuels supply over 80% of the world's energy. Since the energy crises in the 1970's and then in the 90's, when concerns appeared about greenhouse effects, the search for alternative energy sources has been ongoing. Hydrogen has been a particularly popular candidate because its combustion only produces water. Biotechnology is uniquely poised for providing a means for using hydrogen as a source of clean energy. One possibility is using enzymes called hydrogenases that naturally occur in various microorganisms that live in anaerobic ecosystems, such as some bacteria living in soil and in the intestinal tract of animals, or unicellular algae.
Hydrogenases catalyze the conversion of protons in hydrogen molecules (H2), whose combustion releases energy that can be utilized for example in fuel cells and therefore be part of biotechnological devices. The active site that catalyzes this reaction contains metallic ions (Iron or both Iron and Nickel). The Iron-only variety of hydrogenases is the most active for the production of hydrogen molecules. Their remarkably complex active site –the so-called H-cluster– is buried within the core of a large protein. A fatal problem for being able to exploit hydrogenases in biotechnological applications is that when brought to the aerobic conditions of a bioreactor (under normal oxygen pressures), molecular oxygen degrades their active site. Understanding the mechanism of the degradation process of the H-cluster is therefore essential to design a hydrogen-based fuel cell, but studies so far had not been conclusive.
To solve this conundrum, an international team of researchers has combined experiments, molecular simulations, and theoretical calculations. Using electrochemical methods, they have precisely measured the rates of the different reaction steps involved in the degradation of the enzyme by oxygen. They have studied the dependence of these rates on experimental parameters like the electrode potential, pH, H2O/D2O exchange, and mutation of specific amino acids in the protein. These results confirm predictions from theoretical calculations. On one hand molecular dynamics simulations, conducted by nanoGUNE's Ikerbasque Fellow David de Sancho, show the tunnels that oxygen follows to reach the active site of the protein, a necessary step for the degradation and to identify possible hot-spots for blocking these access tunnels. On the other, density functional theory has been used to elucidate the reaction products and evaluate the rate constants for the individual reaction steps.
The study published on August 22nd in Nature Chemistry has allowed to characterize unambiguously the complex reactions that occur in these large biological macromolecules using a highly innovative combination of computational and experimental approaches. "Although important challenges remain ahead for industrial applications, this study opens new avenues to efficiently exploit enzymes from living systems for clean energy production", says De Sancho.
More information: Adam Kubas et al. Mechanism of O2 diffusion and reduction in FeFe hydrogenases, Nature Chemistry (2016). DOI: 10.1038/nchem.2592
Journal information: Nature Chemistry
Provided by Elhuyar Fundazioa