New antiviral drugs could come from DNA 'scrunching'
Evidence of DNA "scrunching" may one day lead to a new class of drugs against viruses, according to a research team from the Perelman School of Medicine at the University of Pennsylvania, the Georgia Institute of Technology, and Columbia University. The team is led by Stephen C. Harvey, PhD, an adjunct professor in the department of Biochemistry and Biophysics at Penn. The scientists report in The Journal of Physical Chemistry B that DNA may go through a repetitive cycle of contraction and elongation, or as they put it, "scrunching," to generate the forces required to drive the DNA into a virus during replication. A better understanding of viral reproduction could be the basis of new ways to fight infectious pathogens.
During replication, viruses take over the host cell machinery to make copies of their genetic material and build protein shells called capsids to contain their genomic DNA or RNA. In some DNA viruses, such as herpesviruses, the empty capsid forms first, and the DNA is then packaged by a protein "motor" on the capsid.
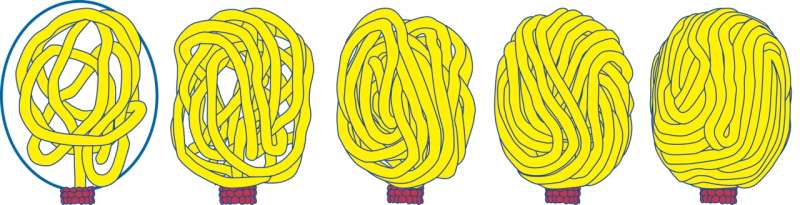
In 2015, Harvey proposed that the traditional model in which the proteins push the DNA forward with a series of lever-like motions might be wrong. He suggested that the proteins might generate a cycle of DNA dehydration and rehydration. DNA is known to shorten by about 25 percent when it is dehydrated. He proposed that the resulting cycle of shortening and lengthening motions could be coupled to a DNA-protein grip-release cycle to generate forward motion. He called this the "scrunchworm model."
"For some time now, we have been contemplating how viral DNA gets into the capsid so that one day we can block this step as a way to halt infection," Harvey said. They tested the scrunchworm model in a series of computer simulations. The structures of the herpesviruses are not known with sufficient resolution to permit this kind of modeling, so the team examined DNA packaging in phi29, a DNA virus of similar structure that infects bacteria. They examined the interaction of DNA with the phi29 connector proteins, which form half of the protein motor. The DNA spontaneously underwent scrunching motions, without being pushed or pulled by protein levers. This provides the first support for the scrunchworm model.
It is essential to test the scrunchworm model experimentally, and Harvey has formed a collaboration with two other research groups to test predictions made by the scrunchworm model. These involve grabbing a single viral particle with a pair of "laser tweezers" and pulling on the DNA tail as the DNA is packaged. The model predicts that DNAs with different sequences will generate different amounts of force, and that DNA with RNA inserts cannot be packaged.
"Even if these experiments disprove the scrunchworm model, they will provide information that will help us figure out how these motors work," Harvey said. "The purpose of modeling is to drive experiments and simulations that advance our understanding, regardless how they turn out."
More information: James T. Waters et al. DNA Scrunching in the Packaging of Viral Genomes, The Journal of Physical Chemistry B (2016). DOI: 10.1021/acs.jpcb.6b02149
Abstract
The motors that drive double-stranded DNA (dsDNA) genomes into viral capsids are among the strongest of all biological motors for which forces have been measured, but it is not known how they generate force. We previously proposed that the DNA is not a passive substrate but that it plays an active role in force generation. This "scrunchworm hypothesis" holds that the motor proteins repeatedly dehydrate and rehydrate the DNA, which then undergoes cyclic shortening and lengthening motions. These are captured by a coupled protein–DNA grip-and-release cycle to rectify the motion and translocate the DNA into the capsid. In this study, we examined the interactions of dsDNA with the dodecameric connector protein of bacteriophage ϕ29, using molecular dynamics simulations on four different DNA sequences, starting from two different conformations (A-DNA and B-DNA). In all four simulations starting with the protein equilibrated with A-DNA in the channel, we observed transitions to a common, metastable, highly scrunched conformation, designated A*. This conformation is very similar to one recently reported by Kumar and Grubmüller in much longer MD simulations on B-DNA docked into the ϕ29 connector. These results are significant for four reasons. First, the scrunched conformations occur spontaneously, without requiring lever-like protein motions often believed to be necessary for DNA translocation. Second, the transition takes place within the connector, providing the location of the putative "dehydrator". Third, the protein has more contacts with one strand of the DNA than with the other; the former was identified in single-molecule laser tweezer experiments as the "load-bearing strand". Finally, the spontaneity of the DNA–protein interaction suggests that it may play a role in the initial docking of DNA in motors like that of T4 that can load and package any sequence.
Journal information: Journal of Physical Chemistry B
Provided by University of Pennsylvania School of Medicine