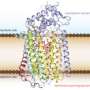
Geobacillus thermodenitrificans. Credit: .hindawi
(Phys.org)—An international team of researchers has revealed for the first time, the structure of an enzyme that pathogenic bacteria use to reduce oxygen. In their paper published in the journal Science, the team describes their study of the atomic structure of a bd oxidase from the bacteria Geobacillus thermodenitrificans, and why they believe what they found might help to create new ways to combat bacterial infections. Gregory Cook with the University of Otago in New Zealand and Robert Poole with the University of Sheffield, in the UK have published a Perspective piece on the work done by the team and why they believe it is important in the study of pathogenic bacteria.
As the researchers note, microorganisms have, over millions of years, evolved a host of enzymes that serve to reduce oxygen to prevent oxidative stress. One such group are oxidases, which Cook and Poole note, fall into three main categories. The first are involved with assisting mitochondria and rely on heme-copper. The second are found in some fungi, plants and certain bacteria and rely on heme-free iron mechanisms that offer nitric oxide resistant respiration. The third case involves bacteria that have cytochrome bd–type oxidases which help to reduce oxygen to water thus avoiding the creation of reactive oxygen species—they have a unique heme composition which prior research has shown is made of two heme b and one heme d, but the exact structure has until now, remained unknown.
In this new effort, the researchers took a closer look at this third category by purifying samples of bd oxidase from G. thermodenitrificans and analyzing its crystal structure. In so doing they found, among other things, that it has 19 helices (CydA and CydB each had nine—the 19th helice was a small peptide called CydS) that spanned the bacterial cytoplasmic membrane and that it was made of up three different heme subunits which are set in a triangular formation (rather than in straight lines as has been found in other types of oxidases) which the team suggests, allows for enzymes to reduce the oxygen present without allowing for the formation of radicals, which can of course, be harmful.
As Cooke and Poole note, this new better understanding of an enzyme used by pathogenic bacteria could help in the development of new, targeted drugs used for treating infections.
More information: S. Safarian et al. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases, Science (2016). DOI: 10.1126/science.aaf2477
Abstract
The cytochrome bd oxidases are terminal oxidases that are present in bacteria and archaea. They reduce molecular oxygen (dioxygen) to water, avoiding the production of reactive oxygen species. In addition to their contribution to the proton motive force, they mediate viability under oxygen-related stress conditions and confer tolerance to nitric oxide, thus contributing to the virulence of pathogenic bacteria. Here we present the atomic structure of the bd oxidase from Geobacillus thermodenitrificans, revealing a pseudosymmetrical subunit fold. The arrangement and order of the heme cofactors support the conclusions from spectroscopic measurements that the cleavage of the dioxygen bond may be mechanistically similar to that in the heme-copper–containing oxidases, even though the structures are completely different.
Journal information: Science
© 2016 Phys.org