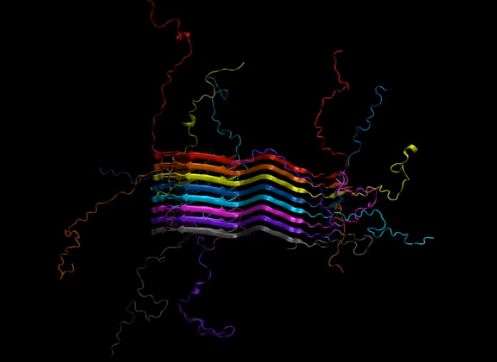
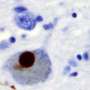
Chemists have identified the complex chemical structure of the protein that stacks together to form fibrils in the brains of Parkinson's disease patients. Armed with this knowledge, researchers can identify specific targets for diagnosis and treatment.
University of Illinois chemists, collaborating with peers at the University of Pennsylvania, Vanderbilt University and Queen Mary University of London, detailed their mapped structure of the protein in the journal Nature Structural and Molecular Biology.
In Parkinson's, the protein alpha-synuclein forms long fibrils that disrupt brain activity. This is similar to the beta-amyloid fibrils that form in Alzheimer's disease patients. However, while the beta-amyloid structure is known, the alpha-synuclein structure has eluded researchers as a result of its complexity, its insolubility and the difficulty of characterizing one protein within a fibril.
"This is the first structure of the full-length fibril protein, which is now well established to be important for the pathology of Parkinson's disease," said study leader Chad Rienstra, a University of Illinois chemistry professor. "Knowing that structure will open up many new areas of investigation for diagnosing and treating Parkinson's disease."
The Illinois group used a special type of molecular imaging called magic-angle spinning nuclear magnetic resonance to measure the placement of atoms in six different samples of alpha-synuclein. In each set of samples, they looked at different sets of atoms, then used advanced computational power to put them all together like pieces of a giant jigsaw puzzle.
Led by University of Illinois professor Chad Rienstra, chemists have identified the full chemical structure of the protein that forms fibrils in the brains of patients with Parkinson’s disease.
"We had to find patterns in the data and systematically test all the possibilities for how the protein would fit together," Rienstra said. "It's like when you solve a really complex puzzle, you know you have it right at the end because all the pieces fit together. That's what we got with this structure."
The group experimentally verified the structure with collaborators by producing the protein in the lab and checking it with various imaging methods to see if it matched the fibrils found in Parkinson's patients. They also verified it biologically by testing it in cell cultures and seeing that it indeed behaved like the protein found in patients.
"These structures are crucial for understanding the mechanisms for how Parkinson's disease works," said Marcus Tuttle, first author of the paper, who worked on the project as a graduate researcher in Rienstra's group and is now a postdoctoral fellow at Yale University. "Amyloid diseases are incredibly complex systems. What structural features drive pathology? That's a super interesting question, but until now there's been no structure. Now there's a whole avenue where we can start to explore the basic mechanism of how the protein works."
Rienstra's group is working with the Michael J. Fox Foundation to identify possible diagnostic agents that could target certain spots on the alpha-synuclein protein and would "light up" in a brain scan, allowing for earlier and more accurate diagnosis.
"We think that the structure that we resolved of alpha-synuclein fibrils will be really significant in the immediate future and has use for diagnosing Parkinson's in patients before they're symptomatic," Rienstra said. "Once people start having symptoms, whether of Alzheimer's or Parkinson's, in many ways it's a little too late to be effective with therapy. But if you catch it early, I think there's a lot of promise for therapies that are being developed. Those are all relying upon the structures that we're solving."
More information: Marcus D Tuttle et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein, Nature Structural & Molecular Biology (2016). DOI: 10.1038/nsmb.3194
Journal information: Nature Structural and Molecular Biology , Nature Structural & Molecular Biology
Provided by University of Illinois at Urbana-Champaign