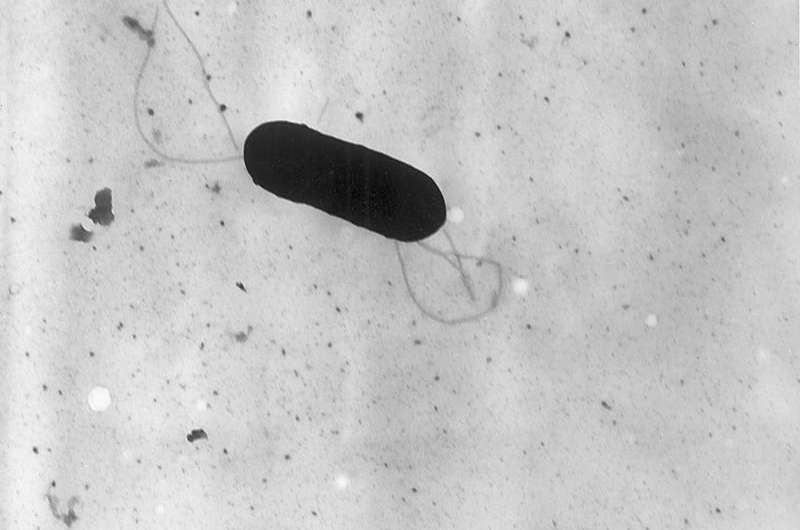
Electron micrograph of a flagellated Listeria monocytogenes bacterium, Magnified 41,250X. Credit: CDC/public domain
(Phys.org)—A large team of researchers affiliated with several institutions in France has conducted a two-pronged study of the bacteria that causes listeriosis and in so doing has identified strains that are more virulent than others and under which circumstances. In their paper published in Nature Genetics, the team discuses their study and outlines their findings.
Listeria is a type of bacteria that infects both humans and other warm blooded animals, it is typically associated with food that is ingested and thus is considered to be a type of food poisoning. Infections can result in flu-like symptoms, eg., pain, cramps, fever, chills, vomiting and headaches, and is considered to be dangerous for older people, pregnant women and those with a compromised immune system. And while much has been learned about the bacteria, there is still a lot that is unknown. In this new effort, the team in France sought to sort out the various strains of the bacteria to learn which are the most infectious and which causes the most problems for those infected.
The researchers took a two-pronged approach to studying the bacteria, the first involved collecting 6,633 samples from various sources and then comparing what they found with already available medical data to ascertain which strains are most likely to infect humans and to cause symptoms, noting also which types were most problematic for those with compromised immune systems.
The second part of the study involved comparing the genomes of 104 common strains of the bacteria to gain a better understanding of the differences in physical makeup between those that are more infectious, or cause more problems in hosts. As part of that study, the team was able to identify a new group of genes that appear to be involved in controlling an individual bacterium's ability to infect the central nervous system and also highlighted previously unknown virulence factors, particularly in the central nervous system or maternal-neonate infection-related clones, which were seen to be particularly virulent in mice.
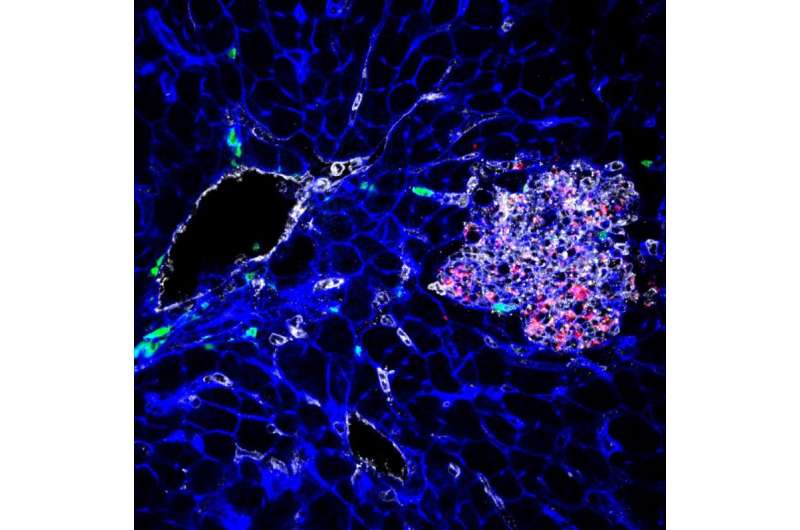
This image shows tissue infected by Listeria (bacteria appear in red). Credit: YH Tsai, M Lecuit, © Institut Pasteur.
The overall result of the study was the identification of the most hypervirulent listeria strains and an improved database holding information regarding Listeria monocytogenes, which health care workers can use to improve treatment for patients—it should also help other researchers looking to better understand the bacteria and ways to prevent people from getting infected.
More information: Mylène M Maury et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity, Nature Genetics (2016). DOI: 10.1038/ng.3501
Abstract
Microbial pathogenesis studies are typically performed with reference strains, thereby overlooking within-species heterogeneity in microbial virulence. Here we integrated human epidemiological and clinical data with bacterial population genomics to harness the biodiversity of the model foodborne pathogen Listeria monocytogenes and decipher the basis of its neural and placental tropisms. Taking advantage of the clonal structure of this bacterial species, we identify clones epidemiologically associated either with food or with human central nervous system (CNS) or maternal-neonatal (MN) listeriosis. The latter clones are also most prevalent in patients without immunosuppressive comorbidities. Strikingly, CNS- and MN-associated clones are hypervirulent in a humanized mouse model of listeriosis. By integrating epidemiological data and comparative genomics, we have uncovered multiple new putative virulence factors and demonstrate experimentally the contribution of the first gene cluster mediating L. monocytogenes neural and placental tropisms. This study illustrates the exceptional power in harnessing microbial biodiversity to identify clinically relevant microbial virulence attributes.
Journal information: Proceedings of the National Academy of Sciences , Nature Genetics
© 2016 Phys.org