February 24, 2016 report
Mapping the nuclear pore complex: 1.5 billion years of innovation
(Phys.org)—If asked to describe the differences between humans and frogs, a child might say that one hops and rib-its while the other walks and talks. If we ask that same child how to build a frog, they will probably need a few minutes with Google. Assuming they are good, they might find that for humans you start with a diameter of 5.2nm for their nuclear pore complexes (NPCs), while for frogs the parameter you use is 10.7nm. That one detail determines a lot about what is possible in the cell, and therefore the entire organism.
In a recent paper published in Plos Biology, researchers from Rockefeller university went fishing for various NPC 'interactomes' in order to trace of the origins and 1.5 billion year evolutionary history of Eukaryotes. The interactome is exactly what it sounds like: any exhaustive set of molecules that can be connected with thin black lines in a figure in a research paper. Practically speaking, this meant starting with a few fluorescently-tagged versions of known core nuclear pore component proteins (called nucleoporins or Nups), and 'walking out' from there using affinity capture and mass spectrometry to identify other proteins that stick.
While there are plenty of contenders for the title of world's greatest protein complex—the ribosome, proteosome, ATPase, and centromere for example—the NPC may be the mightiest of all. At an undisputed 50 MEGADaltons (124 for the mammalian), the NPC contains about 500 subunits comprised of some 30 different Nups. Rather than splashed together like a respiratory complex, the NPC is carefully assembled into an 8-fold symmetric structure that rivets the double nuclear membrane together. To give some idea of the budget that cells are on, the nucleus might have 2000 NPCs (more if mitosis is coming), each ferrying consumables at a rate of 1000 translocations per second.
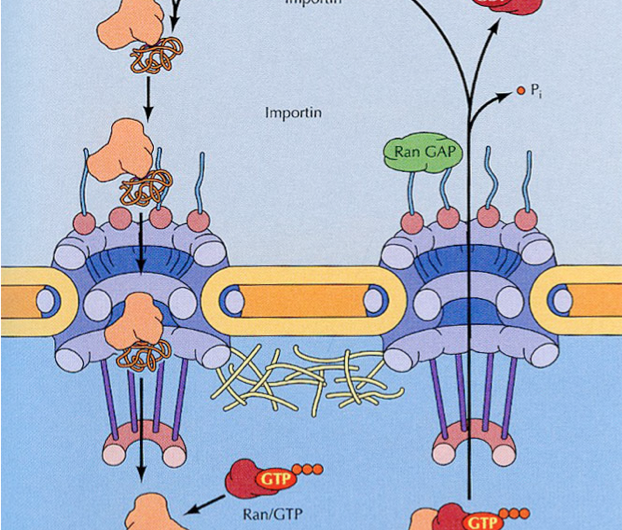
What exactly is a translocation you might ask, and who gets to go in or out? That depends on a lot of things, like that minimum pore size we mentioned above. Although there is no hard and fast size limit to what can pass through by unaided diffusion, things slow down considerably for proteins at around 60kDa. Above that, translocation doesn't become energy dependent per say, but ultimately the ferryman needs to get paid with the hydrolysis of two GTP each time the turnstyle turns. The way it works is neatly described by something known as the Ran-GTP cycle. Many folks are familiar with the idea of gradients across membranes (usually electrical, proton, sodium or other ion), which are harnessed to power auxiliary movements.
The Ran cycle is said to run on a Ran protein gradient where the concentration of the GTP bound form is high inside. and GDP bound form is low outside the nucleus. The tricky part is biasing the pore to get things moving in the right direction. Ribosomes and mRNAs made in the nucleus need to get out, while nuclear proteins translated in the cytoplasm need to get in. These affairs are all neatly enforced by an expansive array of adapters which recognize and bind canonical nucleic acid cytoplasmic localization sequences on the former, and amino acid nuclear localization sequences on the later.
The reason for going into all this detail here is that to understand where Eukaryotes came from we need to understand the partitioning of the cell. The origins of NPCs (and therefore the nucleus) is only half the equation. The other half is mitochondria. The link between the two is established when we realize that in unloading almost all their DNA to the nucleus, the mitochondria had to solve much the same re-localization problem that the NPC translocators have to solve. To get their own protein products, now encoded and made by the host cell, mitochondria developed an analogous (but different) double membrane system of import/export translocators known as the Tim-Tom and Sam families of proteins. In chloroplasts, these are known as the Tic-Toc translocators. The kicker is, if we take Nick Lane's word for it, the NPC first evolved because of the pressures applied by the acquisition of the mitochondria:
Namely, the NPCs served to fence out the ribosomes from the nucleus so that the relatively slow operating spliceosomal machinery had time to work on the many rogue introns and newly acquired endosymbiont genes which managed to get mitochondrial localization sequences appended to themselves.
The remarkable thing that the Rockefeller group found in their studies from the dawn of eukaryotes is that despite retaining a similar composition of important protein motifs and domains, there were gaping architectural dissimilarities between opisthokonts (yeast and vertebrates), and a group of creatures known as trypanosomes. The trypanosomes are known for many things, not least of which is being the parasites responsible for Chagas disease and African sleeping sickness. A key feature is that as a class, they have highly evolved (and degenerate as the case may be) mitochondria. The other peculiar thing about trypanosome mitochondria is their extensive use of RNA editing, typically of uracil. Some of their degenerate mitochondria have lost all their tRNA genes and must import them to translate protein. Other trypanosome mitochondria have lost their entire coding capacity altogether, and serve mainly as organelles to synthesize iron sulfur clusters or other eclectic products.
At the other extreme, many trypanosomes have a huge specialized mitochondria known as a kinetoplast, which services their primary flagellum using a unique system of mini- and maxi- DNA circles. These circle parse and replicate the mtDNA in a highly evolved way not seen in any othr known organism. Incidentally, the name of opisthoconts, (of which we are one) comes from "opistho-" or "behind", and "-kont" meaning flagellum. Most primitive creatures like trypanosomes ("trypano" means "to bore"), actually swim backwards, pulling themselves along by the corkscrew action of their flagellum. Only when things get too crowded, like in thickened blood, do they reverse gears and back out.
Among the critical conserved components that the researchers in all eukaryote NUPs were the major protein folds on the core scaffold Nups lining the main pore. The Nups form the two inner rings which are in turn sandwiched between two outer rings. They contain folds known as α-solenoids and β-barrels or propellers. The importance of these folds is increasingly appreciated as they continue be found at the heart of many newly determined protein crystal structures. The mitochondrial Tom and Sam translocases have them, as do various vesicle coat proteins, including clathrin/adaptin, COPI, and COPII proteins. The authors note that these proteins share architectural characteristics with outer ring Nups, and hint at a common ancestry between the endomembrane trafficking system and the NPC. This so-called 'proto-coatomer hypothesis' further suggests that key components of the cell's secretory system took origin by virtue of their ability to bend membranes, a key first step in the assembly of the pore.
The further one goes out from the core, the less one finds NPC features conserved in the various clades across the millennia. Many peripheral proteins which interact with the core, like the importins and exportins which bind the localization signals and adapters, similarly tend to diverge. This general trend seems to closely parallel the predictable complexification of other major assemblies in the cell. For example, we recently detailed how comparative phylogenetics has unmasked the conserved cores of the ribosome, common enzyme complexes, ion channels, and the GTP coupling so essential to everything from the NPC cycle to olfactory reception.
One specific feature seemingly unique to NPCs the authors found is a loss of an initial symmetry over time: in the ancestral condition it seems that the phenylalanine-glycine (FG) repeats responsible for the selective permeability barrier were fairly uniformly distributed. Over time, creatures from yeasts to man developed a critical asymmetric distribution, favoring FG repeats on either the nuclear or cytoplasmic side. The significance and ubiquity of a tiny feature like this has yet to be seen, however in the protein business, today's anecdote is frequently tomorrow's canonical motif.
More information: Richard Robinson. The Trypanosome Nuclear Pore Reveals 1.5 Billion Years of Similarities and Differences, PLOS Biology (2016). DOI: 10.1371/journal.pbio.1002366
Samson O. Obado et al. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex, PLOS Biology (2016). DOI: 10.1371/journal.pbio.1002365
Journal information: PLoS Biology
© 2016 Phys.org