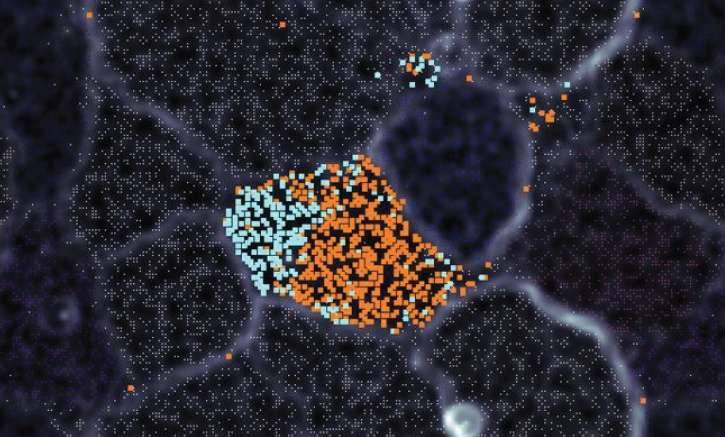
PNNL researchers found a surprising diversity (shown here as blue and orange dots) among genomes within a microbial community, proving that microdiversity plays an important role. This new understanding has important implications for using these communities as a clean energy source.
Tiny microbes are hiding big secrets. Scientists often use a collection or community of microbes to study molecular functions, but the more complex the community, the more difficult it is to tease out functions and interactions. Now, scientists at Pacific Northwest National Laboratory have peered into two microbial communities of blue-green algae to collect near-complete genomic information for all 20 members in each community.
These communities, called unicyanobacterial consortia, or UCCs, are the most complex communities to have their genome described in detail to date. As recently reported in Applied and Environmental Microbiology, the researchers made two surprising discoveries that will help scientists better understand the communities' functions and interactions.
For one thing, the community composition was more varied than typical analyses would have estimated. PNNL's more in-depth analysis showed that usual approaches can underestimate the true diversity and function present within a community. For another, closely related organisms had tiny differences in their genomes. Normally, such microdiversity would put these organisms in competition with each other, and one would drive the other to extinction. Yet these organisms were coexisting in their communities, suggesting they have different functions.
Microdiversity refers to the differences in organisms that have highly similar physical or genetic characteristics. Current thinking assumes that microdiversity in the communities PNNL studied would be minor. However, PNNL's research shows microdiversity is a fundamental property of microbial communities. These differences can have profound impact on understanding how UCCs function, particularly as these communities are being considered for use as a form of clean energy.
"The idea behind this project was to characterize the interactions between organisms-get a complete understanding of how they work together, how they form a community greater than the sum of its parts," said Dr. William Nelson, the PNNL microbiologist who led the study. "Our work shows that these communities are more diverse than anyone expected."
To coax the microbial communities into giving up their secrets, PNNL scientists borrowed a technique called "genome reconstruction" from the environmental field. The relatively new approach allows scientists to segregate species into bins to reconstruct their genomes. Because the same organisms were present in each community at different abundances, the scientists could more easily parse out genomic information.
The team then investigated the function of individual organisms and predicted interactions. To confirm the process's accuracy and specificity, researchers compared the sequenced genomes of isolated organisms from the UCCs. The results appeared fairly accurate, lending support for other scientists to use this method.
"The UCCs are excellent systems from which to learn about how microbial communities behave," Nelson said. "They are much simpler than natural communities, making it easier to interpret what's going on. We have them growing in the lab, so we can perform experiments on them. And, now we have the genome sequences of all the organisms, which gives us a better ability to both make predictions and interpret results."
PNNL scientists are looking into how the small differences in genome sequences translate to function. The UCCs will form the basis for upcoming experiments exploring interactions between community members, including how microdiversity is maintained and how it affects community function. In addition, the results of the current study can serve as a foundational dataset and resource for future investigations to understand interactions among microbial communities, particularly those important to human health and the environment.
More information: William C. Nelson et al. Identification and resolution of microdiversity through metagenomic sequencing of parallel consortia, Applied and Environmental Microbiology (2015). DOI: 10.1128/AEM.02274-15
Journal information: Applied and Environmental Microbiology
Provided by Pacific Northwest National Laboratory