Hierarchical self-assembly of supramolecular muscle-like fibers
The macroscopic movement of our muscles is caused by the collective movement of "biomolecular motors". Scientists and engineers have long been trying to imitate this process. French scientists have now come a good way closer to this goal. In the journal Angewandte Chemie, they have introduced a method for making an artificial muscle that is based on the hierarchical self-assembly of supramolecular muscle-like fibers. Using this technique, they were able to amplify a molecular motion up to the microscopic scale.
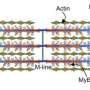
Our muscle tissue has a hierarchical structure: The smallest functional unit of the muscle fibers is called a sarcomere. The sarcomere is made of an actin filament and a myosin filament. The structure of the myosin is similar to a bundle of tiny golf clubs, whose "heads" protrude into the thinner actin filaments. The coordinated motion of thousands of these heads causes the myosin fibers to glide along the actin filaments, which makes the sarcomere contract. Many of these sarcomere units are longitudinally bound to myofibrils that are also laterally bundled into fibers.
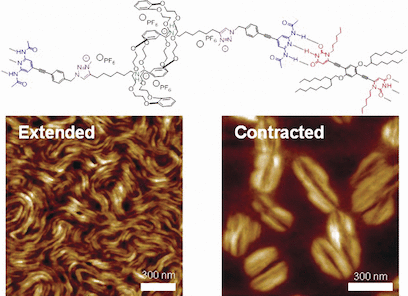
A team headed by Nicolas Giuseppone at the University of Strasbourg and the University of Paris Diderot has now imitated this structural principle. Their individual structural units are rotaxanes, flexible molecular systems made of a rod-like molecule "threaded" through a larger molecular ring. It is possible to make dimers with rotaxanes if a ring is firmly bound to one end of the rod. Within the dimer, the rod of each molecule is threaded through the ring of the other. Such systems have previously used as the basis for molecular switches, because displacement of the rings relative to each other along the axis allows for telescopic contractions and extensions.
The researchers constructed their rotaxane dimers so that the rings each have two "lock down" positions on the axes. In these positions, the rings can be fixed along the axis by means of attractive forces to specific groups of atoms. By switching between acidic and basic conditions the rings can be switched back and forth between the two positions, which changes the length of the dimer in a controlled manner. For the first time, the scientists have been able to link thousands of these dimers into a micrometer-long supramolecular fiber. To do this, they used linking units that bind to special sites at the ends of the dimers by means of hydrogen bonds. This is analogous to the base pairing found in the DNA double helix. The linking units also attract each other (by means of π–π stacking and van der Waals forces), causing the individual fibers to aggregate into bundles that are 10 to 20 nm thick. Electron microscope images showed that when they are contracted, these fibers become thicker like a strained muscle; when they are stretched, they take on an elongated form.
More information: Antoine Goujon et al. Hierarchical Self-Assembly of Supramolecular Muscle-Like Fibers, Angewandte Chemie International Edition (2015). DOI: 10.1002/anie.201509813
Journal information: Angewandte Chemie , Angewandte Chemie International Edition
Provided by Angewandte Chemie