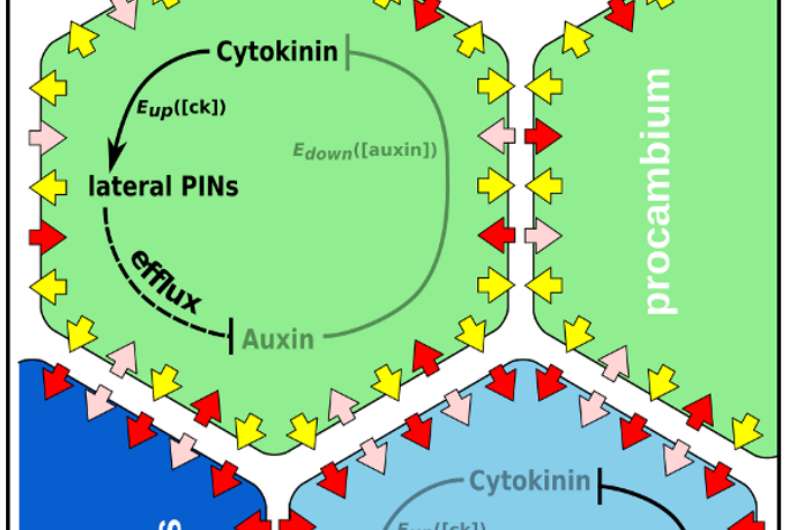
Computational simulations to test the auxin-cytokinin network in a root cross section showed that the network is enough to form the observed patterns in roots. It also identified unexpected limits on cytokinin movement and patterning. Credit: Sedeer el-Showk
Researchers at the University of Helsinki have discovered that cytokinin patterning, an important process in plant development, cannot happen via diffusion alone. While investigating a regulatory network in plant roots, they identified unexpected physical constraints on how cytokinin patterns form.
Interactions between the hormones auxin and cytokinin are central in plant development. In the growing root, cytokinin activates molecular exporters which pump auxin out of cells and auxin activates a gene which blocks cytokinin activity. This mutual inhibition is believed to form an exclusive domain for each hormone. The cells where auxin accumulates become xylem cells, which make up wood.
Sedeer el-Showk, a PhD student at the University of Helsinki, used computational simulations to test the auxin-cytokinin network in a root cross section. The simulations showed that the network is enough to form the observed patterns, but also identified unexpected limits on cytokinin movement and patterning.
Molecular transporters are known to regulate the movement of auxin, but similar transporters have not been found for cytokinin. Nevertheless, a gradient in the distribution of cytokinin is thought to be important in several plant developmental processes, from patterning in the early embryo to continuous growth of the shoot.
However, the simulations showed that cytokinin diffusion cannot form a pattern on a scale small enough for these tissues. "It's easy to propose a cytokinin gradient when you're doing an experiment, but it turns out that physical constraints make it very unlikely that diffusing cytokinin forms a pattern on these scales," explains lead author Sedeer el-Showk. "The pattern has to be created in another way. It could be a pattern in the cytokinin receptors or it might be that there are cytokinin transporters waiting to be discovered. Either way, there's a mystery to solve."
The work was carried out in collaboration with the groups of Verônica Grieneisen and Stan Marée at the John Innes Centre in the UK.
"Our computational work started with experimental data. We've confirmed our simulations with new experiments, and then our computational model has also indicated new lines for experimental work," says group leader Ari Pekka Mähönen. "The back-and-forth between experiments and computational work means the two approaches reinforce and double-check each other."
The study has been published in PLOS Computational Biology.
More information: PLOS Computational Biology, dx.doi.org/10.1371/journal.pcbi.1004450
Journal information: PLoS Computational Biology
Provided by University of Helsinki