September 4, 2015 report
How nature punches back at giant viruses
(Phys.org)—What have viruses ever done for humans? The question is debatable, but given the prevalence of highly contagious, and sometimes life-threatening illnesses caused by viruses, it's fair to say that most people would like to see the tables turned on these often-nasty bundles of DNA strands.
Recently, biologists have become aware of double-stranded DNA virus satellites, which are called virophages because they prey on so-called "giant viruses," or nucleocytoplasmic large DNA viruses (NCLDVs). Now, these NCLDVs don't infect humans; they are parasites of unicellular eukaryotes, but since those are at least in the same taxonomic domain as humans (eukaryota), they can be regarded as the "home team" in a discussion of the misfortunes of giant viruses.
Meet Sputnik
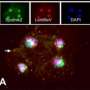
In a study reported in the Proceedings of the National Academy of Sciences, a group of French microbiologists have found around 300 predicted genes of a putative virophage integrated into the nuclear genome of a particular unicellular alga called B. natans. This is evidence that B. natans is a host to NCLDVs. The replication machinery of these viruses is hijacked by virophages, forming a host-virus-virophage tripartite relationship that is the subject of the paper.
The virophage is a satellite virus called Sputnik, which was first described in 2008. It's a member of a new class of icosahedral viruses with a 20 kb, circular, double-stranded DNA genome. It's called a satellite virus because its replication depends on proteins produced by NCLDVs. Sputnik was shown in the previous study to inhibit the replication of its host virus, thus acting as a parasite of that virus. Sometimes pathogenic justice is poetic.
The current study represents the first time microbiologists have uncovered evidence that virophages integrate their DNA into the eukaryotic genome. "This finding led us to predict that B. natans might be prey to NCLDVs," the authors write. "Additional integrated DNA fragments that most probably originate from NCLDV genomes provide data showing that B. natans or its recent ancestor had physical contacts with NCLDV members." The authors hypothesize that the integration of Sputnik's DNA into the eukaryotic host benefits both entities—this symbiosis increases the chances of the virophage to coinfect the cell with its own giant virus prey while simultaneously defending the host cell against a fatal infection.
The integrated virophage genes were highly transcribed, which leads the authors to conclude that they are actually biologically functional; thus, the scenario in which the virophage provides the host with a defense mechanism against NCLDVS. But the authors suggest other possibilities, including the ability of the host to pass on Sputnik's DNA to successive generations: "It is tempting to speculate that integrated virophages can act as molecular weapons against viral pathogens, conferring a sort of immunity transmissible from generation to generation," they write.
The study of virophages is quite new, and the microbiology field has only just begun documenting their characteristics. The authors emphasize that the prevalence and biological significance of virophage DNA insertion will remain open questions until more genomes of putative eukaryotic hosts have been sequenced.
More information: "Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses." PNAS 2015 ; published ahead of print August 24, 2015, DOI: 10.1073/pnas.1506469112
Abstract
Virophages are recently discovered double-stranded DNA virus satellites that prey on giant viruses (nucleocytoplasmic large DNA viruses; NCLDVs), which are themselves parasites of unicellular eukaryotes. This coupled parasitism can result in the indirect control of eukaryotic cell mortality by virophages. However, the details of such tripartite relationships remain largely unexplored. We have discovered ∼300 predicted genes of putative virophage origin in the nuclear genome of the unicellular alga Bigelowiella natans. Physical clustering of these genes indicates that virophage genomes are integrated into the B. natans genome. Virophage inserts show high levels of similarity and synteny between each other, indicating that they are closely related. Virophage genes are transcribed not only in the sequenced B. natans strain but also in other Bigelowiella isolates, suggesting that transcriptionally active virophage inserts are widespread in Bigelowiella populations. Evidence that B. natans is also a host to NCLDV members is provided by the identification of NCLDV inserts in its genome. These putative large DNA viruses may be infected by B. natans virophages. We also identify four repeated elements sharing structural and genetic similarities with transpovirons—a class of mobile elements first discovered in giant viruses—that were probably independently inserted in the B. natans genome. We argue that endogenized provirophages may be beneficial to both the virophage and B. natans by (i) increasing the chances for the virophage to coinfect the host cell with an NCLDV prey and (ii) defending the host cell against fatal NCLDV infections.
Journal information: Proceedings of the National Academy of Sciences
© 2015 Phys.org