Analysis of horse pathogen sheds light on persistent infections
Analysis of an ancient pathogen of horses shows that it went through a major population replacement during the global conflicts of the late 19th and early 20th centuries and has shed light on the genetics of low diversity and persistent infections such as human immunodeficiency virus (HIV) and tuberculosis.
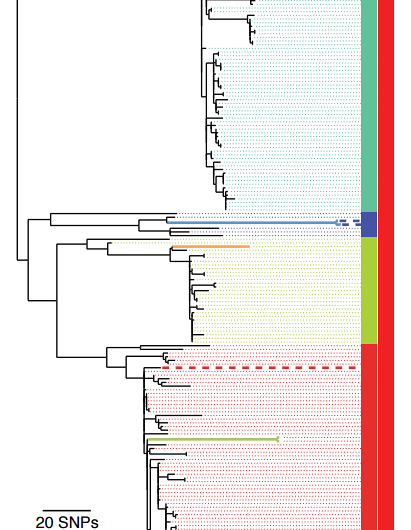
In the largest-ever study of the bacteria Streptococcus equi, which causes the disease strangles in horses, researchers were surprised by the genetic similarity of the 224 samples they had procured from horses across the globe.
The disease was first recorded in 1251, so scientists would have expected to see a number of distinct bacterial lineages that had diversified over the centuries. Looking back through the genomes to reveal the common ancestor, researchers instead identified a population replacement had occurred at the turn of the 20th century, a period when horses from around the world were brought together and suffered high mortality rates in a series of global conflicts including World War I, where an estimated eight million horses died on the battlefield.
"The mobilisation and mixing of horses in conflicts such as WWI provided perfect conditions for Streptococcus equi to thrive," says Simon Harris, first author from the Wellcome Trust Sanger Institute. "This combined with the high mortality rates among the horses and the subsequent intense breeding that produced a new naïve host population could explain why we see so little diversity in modern Streptococcus equi strains."
While loss of diversity could be considered detrimental to bacterial populations, Streptococcus equi still infects more than 20,000 horses in the UK alone each year. Its success, researchers believe, is due to the evolutionary niche it has carved out for itself in horses, where it first causes an acute infection and then persists in the horse for years while showing no visible signs.
Researchers observed gene decay in the persistent samples they sequenced; this is where bacteria shed genes that are no longer useful. In the case of Streptococcus equi living in one horse for long periods, genes that helped the bacteria spread to new hosts and share genetic material with other bacteria became less useful. At the same time, Streptococcus equi adapted to its environment in horses, becoming able to evade the immune system so effectively that more than one strain can infect the same horse and the persistent bacteria is able to resurface, causing multiple periods of painful disease.
"Strangles is a distressing disease that causes large abscesses to form in a horse's head and neck," explains Andrew Waller, Head of Bacteriology at the Animal Health Trust. "The data we have gathered in this study will enable us to pinpoint the genes that help the bacteria to persist, spread and thrive in the horse population. Our hope is that these genes will be effective vaccine targets."
The ability of Streptococcus equi to adapt to living in a persistent state within its host and still infect new horses mirrors the situation with HIV. Researchers hypothesise that in both cases the original genetic sequence that infected the host is more often passed on to the next host. The bacteria or viruses with these ancestral DNA sequences then change and adapt to persist in the new host. This, scientists believe is why diversity appears so low in these populations.
Further parallels can be drawn between Streptococcus equi and the bacteria that causes tuberculosis. The population of bacteria responsible for this respiratory disease in humans also has low diversity between patients but is able to quickly develop antibiotic resistance within its host.
"Unravelling the complex population dynamics of Streptococcus equi sheds new light on the balancing act between acute and persistent infection that is going on in many pathogens," says Matthew Holden, a senior author from the Wellcome Trust Sanger Institute and the University of St Andrews. "Not only does this collection of whole-genome sequences for Streptococcus equi offer hope for an effective strangles vaccine, it also provides us with a useful model for understanding persistent infection in humans."
"The Horserace Betting Levy Board is delighted to have supported this important work," says Annie Dodd, Grants Manager at the Horserace Betting Levy Board. "Streptocuccus equi is a persistent challenge to the horse population worldwide and this significant progress towards an effective vaccine is extremely good news. We are pleased too that this programme may also have useful implications for such devastating human diseases as HIV and TB. We look forward to seeing what happens next."
More information: Genome Research 2015. DOI: 10.1101/gr.189803.115
Journal information: Genome Research
Provided by Wellcome Trust Sanger Institute