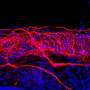
Green fluorescence marks the position of tagged proteins attached to anchors lodged within the membrane of a vesicle. Credit: Andrew Rudd, UC San Diego
With a tag, an anchor and a cage that can be unlocked with light, chemists have devised a simple, modular system that can locate proteins at the membrane of a cell.
"If you're trying to emulate the way nature does this, you need a lot of complex machinery," said Andrew Rudd, a graduate student in chemistry and biochemistry at the University of California, San Diego.
Rudd sought something simpler. He works with Neal Devaraj, an assistant professor of chemistry and biochemistry whose group has been working toward the creation of artificial cells from scratch in part by finding minimal ways to create biological structures.
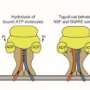
They fused proteins to molecules called SNAP-tags, modified enzymes that recognize a particular chemical group called a benzylguanine. Researchers use SNAP-tags to attach things to proteins, such as fluroescent bits that allow them to find and follow the molecules within a cell. In this case the researchers used SNAP-tags to direct the proteins' movements.
By linking a benzylguanine to a phospholipid, the molecules that make up membranes, the group lodged an anchor that a SNAP-tag would recognize in an artificial membrane. To prevent SNAP-tags from recognizing the anchor right away, they created a chemical cage to protect the benzylguanine.
Tiny beams of ultraviolet light release the cages, allowing SNAP-tag-fused proteins to attach to the membrane anchors.
"What's cool is that we can do this on a very fine scale," Rudd said, with precise control of both timing and location.
Rudd, undergraduate student Joan Valls Cuevas, and Devaraj reported their success in the Journal of the American Chemical Society.
Journal information: Journal of the American Chemical Society
Provided by University of California - San Diego