A greasy way to take better protein snapshots
Thanks to research performed at RIKEN's SACLA X-ray free electron laser facility in Japan, the dream of analyzing the structure of large, hard-to-crystallize proteins and other bio molecules has come one step closer to reality. In the study published in Nature Methods, researchers used a newly developed grease to suspend small crystals of lysozyme, glucose isomerase, thaumatin, and fatty acid-binding protein type-3, which they then analyzed using the revolutionary serial femtosecond crystallography method.
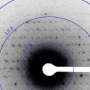
Crystallography, which was first performed just a century ago, has allowed us to understand the structure of molecules, giving us much greater understanding of how life works. However, x-ray crystallography was plagued in the past by the fact that ironically, the x-ray pulses that are used to produce the diffraction patterns which are then analyzed destroy the very molecules that they are analyzing.
This limitation was eventually overcome by the development of XFEL lasers like SACLA, which create such blindingly bright and short—on the order of femtoseconds—pulses of light that they allow the material to be analyzed before it has time to be destroyed through the active of reactive species. This means that the crystals can be much smaller than the bulky samples that had to be created in the past to withstand the onslaught of x-ray beams.
However, the difficulty with the new femtosecond method is that each crystal is damaged as it passes through the beam, so a series of diffraction patterns needs to be taken—of one crystal structure after another—to allow the patterns to be combined to make a good image. In the past, this was generally done by shooting the crystals in a liquid jet through the laser beam, with some being hit and most of them passing through untouched. But this was very inefficient, with on average just one out of a thousand crystals being hit by the beam.
To overcome this, the research team from RIKEN, Osaka University, Kyoto University and the Japan Synchrotron Radiation Research Institute came up with an ingenious idea?suspend the crystals in a greasy substance and then pass the grease through the beam to get the diffraction pattern.
According to Michihiro Sugahara, the first author of the paper, "There were many difficulties involved in this research, but it was a great feeling when we were finally able to come up with precise diffraction patterns of the crystals, using much smaller samples than in previous studies. We were able to collect eight thousand to thirty thousand indexable diffraction patterns in just an hour, using a small sample of less than 1 milligram, and these were used to give us a precise structure of the molecules."
"We hope," he continues, "that this technique could be used for the analysis of dangerous proteins—for example those containing mercury?since the grease holds them in place unless liquid jets, which allow them to escape into the environment."
Martin Weik, from the Institute of Structural Biology in Grenoble, France, says "This is a wonderful development that we have already made use of at SACLA to solve the structure of one of our proteins of interest. In the future, we hope to employ the newly developed grease method to structurally observe our protein at work with femtosecond time resolution."
More information: Grease matrix as a versatile carrier of proteins for serial crystallography, Nature Methods, DOI: 10.1038/nmeth.3172
Journal information: Nature Methods
Provided by RIKEN