Mitochondrial Nucleoid Core. Credit: mol-biol4masters.masters.grkraj.org
(Phys.org) —In the brave new world of three-parent embryos several inherited mitochondrial diseases can potentially be solved. One slightly dubious argument used by its champions to assuage equally dubious traditional ethical objections is that a mitochondrial donor only supplies 16.6K base pairs (BPs) of mtDNA to the child—a trifling amount compared the 3.4B BPs of (nuclear) nucDNA. What this unassailable yet simplistic truth conceals is that with perhaps 10 plasmid copies per mitochondria, and 100K mitochondria per egg, we are actually talking about 16.5B BPs of mtDNA stashed in strategic fortifications throughtout the cell. Although there is considerable redundancy even in cells with a fairly heteroplasmic stock of mitochondria, that's a bit more DNA than the nucleus has.
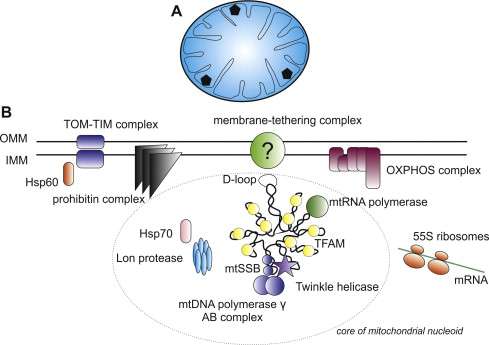
A detailed picture of the structure of the eukaryotic nucleus and the chromosomes within it still eludes modern day cell biology. It is no wonder that a Youtube search can not fulfill that request—we don't even have the video yet for the mitochondrion's plasmid nucleoid. As in many things biological, the best way to try to understand these nucleoids is to build them. In other words reconstitute them in vitro from minimal components. A new paper in Cell Reports describes the construction of mitochondrial nucleoprotein complexes from scratch. By fine-tuning the concentration of a ubiquituous compaction protein known as mitochondrial transcription factor A (TFAM), the researchers demonstrate the precise regulation of mtDNA replication and transcription.
So what might we expect from the definitive mitochondrial animation? Corresponding author Maria Falkenberg would probably be one of the best consultants to have on the project. Her group previously created a minimal mtDNA "replisome" in vitro and established that Twinkle is the helicase used at the mtDNA replication fork. Acting alone, the definitive mtDNA polymerase (POLγ) cannot use double-stranded DNA template for DNA synthesis. However in combination with Twinkle, single strands of DNA up to 2kb can be synthesized. When ssDNA-binding protein (mtSSB) is added to mix, DNA products up to about 16 kb can be made—ie. the same size as mammalian mtDNA.
Using the replisome trinity just described, the researchers can titrate in fluorescently-labelled TFAM and get a hands on feel for the effects using a combination of optical trapping and atomic force microscopy. They found that high TFAM:mtDNA ratios resulted in the formation of large stable TFAM filaments which compact the DNA and reduce the progression of replication and transcription complexes. Small changes in the TFAM concentration generally resulted in a rather large impact on the average compaction.
As in the DNA binding of histones in the nucleus, there remains some ambiguity in the works and the picture is still incomplete. For example, TFAM binds in a cooperative manner (in the sense of cooperative hemoglobin binding to oxygen) to nonspecific DNA sequences forming protein patches with each monomer covering approximately 30bp of DNA. On the other hand, acting as a transcription factor, there appears to be some more specific DNA promoter binding as well, with the establishment of a specific U-turn motiff. Previous in vivo estimates have put the concentration of TFAM somewhere in the range one molecule per 15 to 18 bp of mtDNA.
The compaction is belived to result from a partial unwinding of duplex DNA. This "softening", if you will, effects the higher order structure of the DNA—what DNA topologists call the twist and writhe. Twist is the number of helical turns in the DNA while the writhe is the number of times the double helix crosses over on itself. In mtDNA (and many other types of DNA as well) the writhe component is negatively supercoiled, or slightly unwound in the normal state.
In the statements above regarding the three parent embryos and the estimates of total mtDNA per cell, the assumption was made that each nucleoid contains just a single copy of the 16 kbp molecule. While there is some evidence for that, the full dynamic nature of nucleoids (and mitochondria in general) has yet to be explored. A few years ago, Nick Lane raised concerns about potential mtDNA incompatibilities for three parent embryos. He and others have noted that our genes show all the cardinal signs of compatibility, even optimality, with our mitochondria at the individual level.
This new work on the the nature of mtDNA is critical to our understanding what really controls the cell—and therefore in the case of germ cells—what controls us. Nowhere of course, does it say we must only produce optimal children, or for that matter, healthy ones. More important for us, is to be able to decribe what those terms might even mean.
Comparing brains or cells to computers is a tired analogy. None the less we might tap it for incite into why our mitochondria retain the genes that they have. It is notable that as computer architectures continue to evolve to entirely new problems, hardware concepts are often completely rethought. To defeat a human at Jeopardy, IBM's Watson for example, replicates the mere 4TB of data in its filesystem several times across 16TB of RAM. The expansive replication of select mtDNA throughout the cell likely has similar advantages for life.
More information: In Vitro-Reconstituted Nucleoids Can Block Mitochondrial DNA Replication and Transcription, Cell Reports, dx.doi.org/10.1016/j.celrep.2014.05.046
Journal information: Cell Reports
© 2014 Phys.org