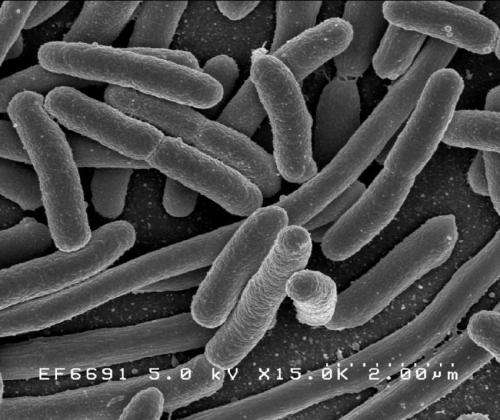
Escherichia coli. Credit: Rocky Mountain Laboratories, NIAID, NIH
(Phys.org) —A team of researchers working at the Hebrew University in Israel has found that Escherichia coli grown in a lab were able to change their dormancy period to match the time that antibiotics were administered—saving themselves from being killed by it. In their paper published in the journal Nature, the team describes how they subjected E. coli to various periods of antibiotic dosing and how the bacteria responded.
There has been a lot of news of late sounding the alarm about growing resistance in harmful bacteria, leading to a time when we will no longer have any drugs to fight infections. In this new study, the news appears to grow worse as the researchers in Israel have found that harmful bacteria such as E. coli are also able to modify their dormancy periods to withstand doses of antibiotics that would normally kill them—an ability known as tolerance.
When bacteria are dormant, they essentially go to sleep—instead of growing, they simply sit idle. While in this state, antibiotics aren't able to kill them because they won't be absorbed. Scientists have known about this for quite some time. What's new is that it appears that some bacteria can modify their dormancy period to coincide with the duration of administration of antibiotics.
To better understand dormancy periods with E. coli, the researchers subject E. coli specimens to a round of ampicillin, which killed approximately 99.9 percent of those present. Those that lived were then allowed to repopulate at which point they received another round of the antibiotic. The researchers repeated this exercise several times and found that those that lived, were able to do so because they were in a dormant state, and surprisingly, were able to maintain their dormant state for the same length of time as the antibiotic dosing period. Further investigation revealed that if the duration of the ampicillin dose was changed, the duration of the bacterial dormant state would change just as much—the bacteria were able to alter their dormancy period after just a few generations (as few as 10 cycles) to match the dosage periods. By doing so E. coli managed to increase its survival rate one hundred fold.
Genetic analysis revealed three genes that appear to play a role in dormancy period alteration, but the actual mechanism behind the ability of the bacteria to change so rapidly is still a mystery.
More information: Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations, Nature (2014) DOI: 10.1038/nature13469
Abstract
The great therapeutic achievements of antibiotics have been dramatically undercut by the evolution of bacterial strategies that overcome antibiotic stress. These strategies fall into two classes. 'Resistance' makes it possible for a microorganism to grow in the constant presence of the antibiotic, provided that the concentration of the antibiotic is not too high. 'Tolerance' allows a microorganism to survive antibiotic treatment, even at high antibiotic concentrations, as long as the duration of the treatment is limited. Although both resistance and tolerance are important reasons for the failure of antibiotic treatments, the evolution of resistance is much better understood than that of tolerance. Here we followed the evolution of bacterial populations under intermittent exposure to the high concentrations of antibiotics used in the clinic and characterized the evolved strains in terms of both resistance and tolerance. We found that all strains adapted by specific genetic mutations, which became fixed in the evolved populations. By monitoring the phenotypic changes at the population and single-cell levels, we found that the first adaptive change to antibiotic stress was the development of tolerance through a major adjustment in the single-cell lag-time distribution, without a change in resistance. Strikingly, we found that the lag time of bacteria before regrowth was optimized to match the duration of the antibiotic-exposure interval. Whole genome sequencing of the evolved strains and restoration of the wild-type alleles allowed us to identify target genes involved in this antibiotic-driven phenotype: 'tolerance by lag' (tbl). Better understanding of lag-time evolution as a key determinant of the survival of bacterial populations under high antibiotic concentrations could lead to new approaches to impeding the evolution of antibiotic resistance.
Journal information: Nature
© 2014 Phys.org