New DNA cleavage technique could lead to more versatile genetic engineering

Genetic engineering of plants, animals and microorganisms such as bacteria typically involves the use of restriction enzymes to 'cut and paste' DNA fragments into certain genetic sequence locations. This process allows scientists to introduce new genes into an organism, but is constrained to specific recognition sequences, limiting the design of recombinant DNA molecules.
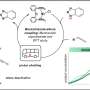
A research team led by Hiroki Ueda and colleagues from the Laboratory for Synthetic Biology at the RIKEN Quantitative Biology Center has now developed a chemical-based, non-enzymatic recombination technique that instead uses a DNA base analogue called 5-ethynyluracil to cleave DNA at any site containing the nucleotide thymine.
The technique developed by Ueda and his co-workers, which is called quantitative base-induced DNA cleavage (QBIC), starts with the generation of DNA fragments containing 5-ethynyluracil in place of thymine—two molecules with similar structures. These products are then immersed in an aqueous solution containing methylamine, a derivative of ammonia. In this chemical bath, all the nucleotides containing 5-ethynyluracil become cleaved, introducing gaps near the cleaved ends. The gaps in the resulting DNA fragments create protruding ends that can be inserted into circular DNA molecules known as plasmids. The plasmids can then be inserted into the target organism, such as a bacterial cell, to complete the genetic engineering process.
"Compared with restriction enzymes, the QBIC reaction has the advantage that we can freely design the sequences at the protruding termini generated by the DNA cleavage," says Katsuhiko Matsumoto from the research team. "The experimental procedure for DNA concatenation using the QBIC reaction is also simple," he adds. "DNA can be concatenated by the addition and removal of methylamine, hybridized by heating and cooling, and incorporated into an organism—in this case the bacterium Escherichia coli."
Another potential boon of the QBIC method is that it is less sensitive to laboratory conditions than enzyme-based techniques and can be run at room temperature. Being a chemical method, it is also generally cheaper to perform than enzyme-based methods. One limitation of the QBIC method in its present form is that long stretches of DNA can lose their structure after treatment with the methylamine solution, which prevents the two-stranded, helical shape from being restored. Ueda's team is now refining the protocol to extend its ability to handle longer DNA fragments. "If we find a solution to this problem," Matsumoto notes, "the QBIC method would become very attractive for the concatenation of long DNA fragments."
More information: Ikeda, S., Tainaka, K., Matsumoto, K., Shinohara, Y., Ode, K. L., Susaki, E. A. & Ueda, H. R. "Non-enzymatic DNA cleavage reaction induced by 5-ethynyluracil in methylamine aqueous solution and application to DNA concatenation." PLoS ONE 9, e92369 (2014). DOI: 10.1371/journal.pone.0092369
Journal information: PLoS ONE
Provided by RIKEN