Inner workings of a cellular nanomotor revealed
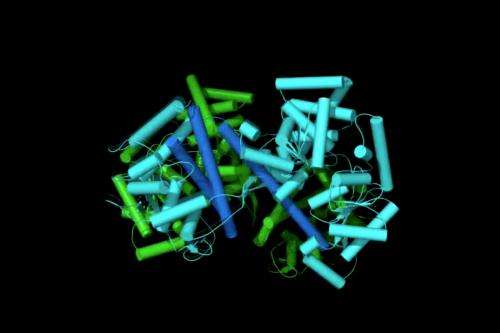
Our cells produce thousands of proteins but more than one-third of these proteins can fulfill their function only after migrating to the outside of the cell. While it is known that protein migration occurs with the help of various 'nanomotors' that push proteins out of the cell, little is known about their precise mechanical functioning. New research by Anastassios Economou (KU Leuven) and his team reveals the inner workings of one such nanomotor, called SecA, with new clarity.
Protein migration is a fundamental problem in biology and is essential for life. Examples of migrating proteins include insulin (the absence of which leads to diabetes), antibodies (essential for combating infections), membrane channels (essential for neuronal cell function) and toxin-proteins (secreted by pathogenic microorganisms).
Migrating proteins contain chemical signals called signal peptides. These signal peptides act as postal addresses and direct exported proteins to the membrane for transport outside the cell.
In previous research, Dr Economou, in collaboration with Babis Kalodimos (Rutgers University), revealed how signal peptides bind to a specific cellular receptor on the membrane, which subsequently connects to the export channel leading out of the cell. This receptor was also found to act as a nanomotor, with two separate piston-like mechanical parts that somehow push proteins out of the cell. The exact mechanism by which this occurred remained a mystery. Until now.
In the current study, published in the December issue of Molecular Cell, Economou and his team show how the SecA receptor moves to push proteins out of the cell: when a signal peptide makes contact, the two pistons become excited. They position themselves one against the other in a series of defined steps and modify their shape. This finely orchestrated series of motions opens the export channel and traps the exported protein inside it. In a final step, the two parts dissociate and the remaining single piston pushes the protein out in cycles of repeated motions.
The discovery adds a significant piece in the puzzle of exploiting protein migration to improve health. "Moving forward, this discovery will help us focus efforts to find specific antibiotics against harmful bacterial protein secretion pathways," says Dr Economou. "It also offers the possibility to optimise biotechnological production of human biopharmaceuticals by using microbial 'cell factories' for secreting biopharmaceuticals. We will be pursuing these avenues in future research."
More information: The paper "Quaternary Dynamics of the SecA Motor Drive Translocase Catalysis" by Giorgos Gouridis, Spyridoula Karamanou, Marios Frantzeskos Sardis, Martin Alexander Schärer, Guido Capitani and Anastassios Economou can be accessed on the website of Molecular Cell: www.cell.com/molecular-cell/ab … 1097-2765(13)00860-5
Journal information: Molecular Cell
Provided by KU Leuven