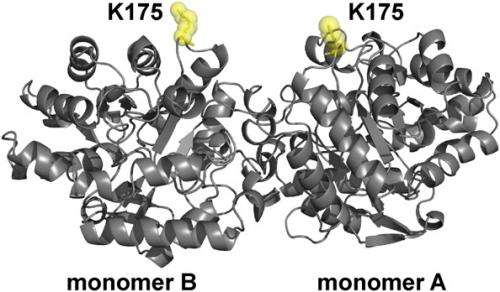
Structure of OPH showing the position of site-specific donor and acceptor labeling. OPH is a homodimer (C2 symmetry) that consists of two (α/β)8 monomers. The position K175, which was replaced with AzF in monomers A and B of OPH, is highlighted (yellow). Credit: Copyright © PNAS, doi:10.1073/pnas.1311761110
(Phys.org) —Proteins accomplish something rather amazing: A protein can have many functions, with a given function being determined by the way they fold into a specific three-dimensional geometry, or conformations. Moreover, the structural transitions form one conformation to another is reversible. However, while these dynamics affect protein conformation and therefore function, and so are critical to a wide range of areas, methods for understanding how proteins behave near surfaces, which is complicated by protein and surface heterogeneities, has remained elusive. Recently, however, scientists at University of Colorado utilized a method known as Single-Molecule Förster Resonance Energy Transfer (SM-FRET) tracking to monitor dynamic changes in protein structure and interfacial behavior on surfaces by single-molecule Förster resonance energy transfer, allowing them to explicate changes in protein structure at the single-molecule level. (SM-FRET describes energy transfer between two chromophores – molecular components that determine its color.) In addition, the researchers state that their approach is suitable for studying virtually any protein, thereby providing a framework for developing surfaces and surface modifications with improved biocompatibility.
Prof. Joel L. Kaar discussed the paper he and his co-authors, Dr. Sean Yu McLoughlin, Prof. Mark Kastantin and Prof. Daniel K. Schwartz, recently published in Proceedings of the National Academy of Sciences. "The primary challenges in devising our approach to characterizing changes in protein structure were implementing a site-specific labeling method, which enabled single-molecule resolution, as well as a method to only image molecules at the solution-surface interface," Kaar tells Phys.org. The scientists overcame the former challenge by incorporating unnatural amino acids – that is, those not among the 20 so-called standard amino acids – with unique functional groups for labeling with fluorophores (chemical compounds that can re-emit light upon light excitation); the latter, by using total internal reflection fluorescence microscopy, which only excites molecules in the near-surface environment, thereby minimizing the background fluorescence of molecules free in solution. "Although site-specific labeling methods have been used to monitor changes in protein conformation mainly in bulk solution, such techniques have not previously been exploited to study freely diffusible protein molecules at interfaces," Kaar adds. As such, the researchers are the first to apply site-specific labeling methods to study protein-surface interactions,
"The major challenge associated with incorporating unnatural amino acids for labeling was related to the optimization of protein expression," Kaar continues. Specifically, he explains, the expression of the enzyme organophosphorus hydrolase (OPH) – which is notoriously difficult to make in large quantities due to inclusion body formation – with the unnatural amino acid p-azido-L-Phe (AzF) had to be optimized to efficiently incorporate p-azido-L-Phe. (Inclusion body formation refers to the intracellular aggregation of partially folded expressed proteins,) "This process required modification of expression conditions," he adds, "in which bacteria with modified genetic machinery were grown to enable production of soluble enzyme for single-molecule experiments."
Moreover, Kaar continues, monitoring molecule-by-molecule structure changes in organophosphorus hydrolase had its own challenges related to eliminating mislabeled protein molecules – that is, molecules with other than one donor and one acceptor fluorophore – from analysis. "We met this challenge by creating and implementing filters during data analysis that separated signals from properly labeled and mislabeled species."
Kaar points out that using SM-FRET tracking had its own issue. For one, it required high-throughput tracking algorithms (developed by co-authors Kastantin and Schwartz) critical to monitor changes in FRET signals for large numbers of molecules, which in turn was essential to identifying protein structure changes accurately (that is, with high statistical confidence). He points out that SM-FRET also required prior knowledge of the crystal structure of OPH, which was needed to make the FRET signal indicative of quantitative changes in protein conformation.
The study's results suggest that surfaces may act as a source of unfolded (that is, aggregation-prone) protein back into solution – but validating this implication faces the challenge of identifying the conformation of protein molecules immediately before desorption from the surface. "The question of whether the unfolded proteins induced aggregation in solution after desorption remains to be fully understood," Kaar explains. "Fully understanding if this is actually the case requires further analysis of protein in solution in the presence of the surface."
The team leveraged two key innovations to address these research challenges – the implementation of site-specific labeling methods, and high-throughput tracking algorithms with SM-FRET. "Combining these methods enabled the decoupling of surface-induced conformational changes from protein adsorption and desorption events," Kaar notes. "By decoupling such phenomenon, this approach allowed us to overcome the limitations of conventional surface characterization methods."
The research also shows that SM-FRET permits a unique level of understanding of the ways in which surface chemistry influences molecular conformation and, in turn, function. "By observing molecular-level changes in protein structure in isolation from competing surface dynamics, it's easier to make a direct connection between surface chemistry and conformation," Kaar points out. "Therefore, it is more straightforward to see the effects of surface chemistry and can lead to new ideas for how to improve chemistry for a given application.
Another important finding is that the new method will enable the creation of surfaces and modifications with improved biocompatibility by uncovering the connection between surface properties and protein unfolding. "This connection is critical to inspiring and developing surfaces and modifications that meld with the biological world," Kaar explains. "For example, with this understanding, we can begin to design surfaces that promote protein folding and therefore favorable responses from cells present in the surrounding milieu. In this example, the folded state of the protein may display certain biological signals to cells that thwart unwanted inflammatory or harmful reactions while instructing cells to respond in ways that may facilitate proliferation, differentiation or even wound healing in vivo."
Kaar tells Phys.org that future experiments are aimed at determining if the observed effects of fused silica on organophosphorus hydrolase are general or specific to this combination of surface and protein. "We plan to address this question by probing how fused silica and surfaces with other properties impact the folding of other proteins. We're also interested in expanding our methods to understand how surface effects on conformation impact the binding of a third protein species. Understanding this impact is critical to, for example, enumerating how cells respond to biological cues on surfaces in physiological environments." Other innovations that the researchers may develop, Kaar adds, include more sophisticated labeling to minimize SM-FRET protein mislabeling on surfaces, as well as labeling and detection schemes to enable multiple molecular events, including unfolding and binding, to be monitored simultaneously.
"Given that the interaction of proteins and surfaces are relevant in virtually all areas of biotechnology," Kaar notes, "many other areas of research – for example, tissue engineering and regenerative medicine, biosensing, biocatalysis, and pharmaceutical protein formulation – may benefit from exploiting our approach."
More information: Single-molecule resolution of protein structure and interfacial dynamics on biomaterial surfaces, PNAS November 26, 2013 vol. 110 no. 48 19396-19401, doi:10.1073/pnas.1311761110
Journal information: Proceedings of the National Academy of Sciences
© 2013 Phys.org. All rights reserved.